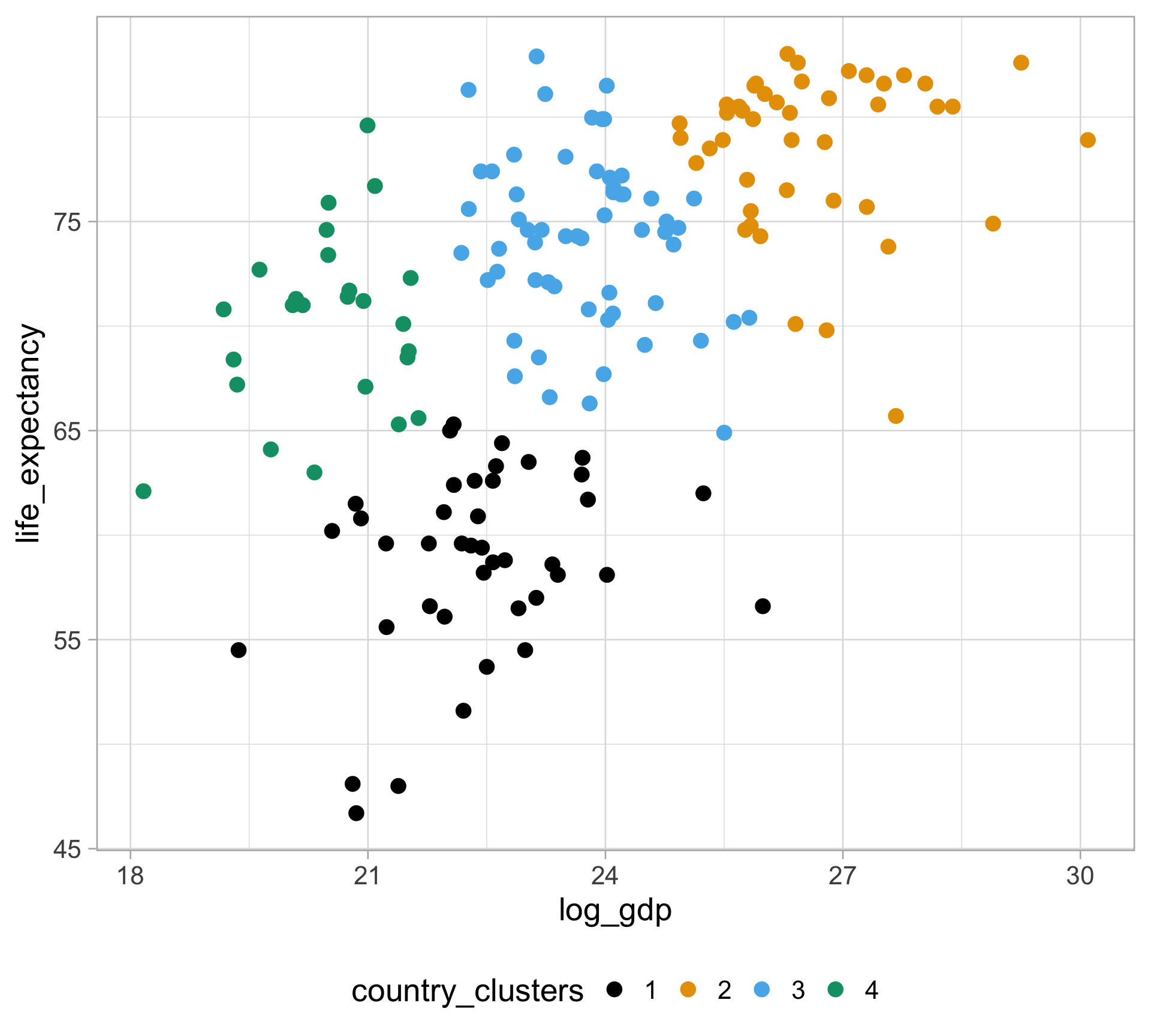
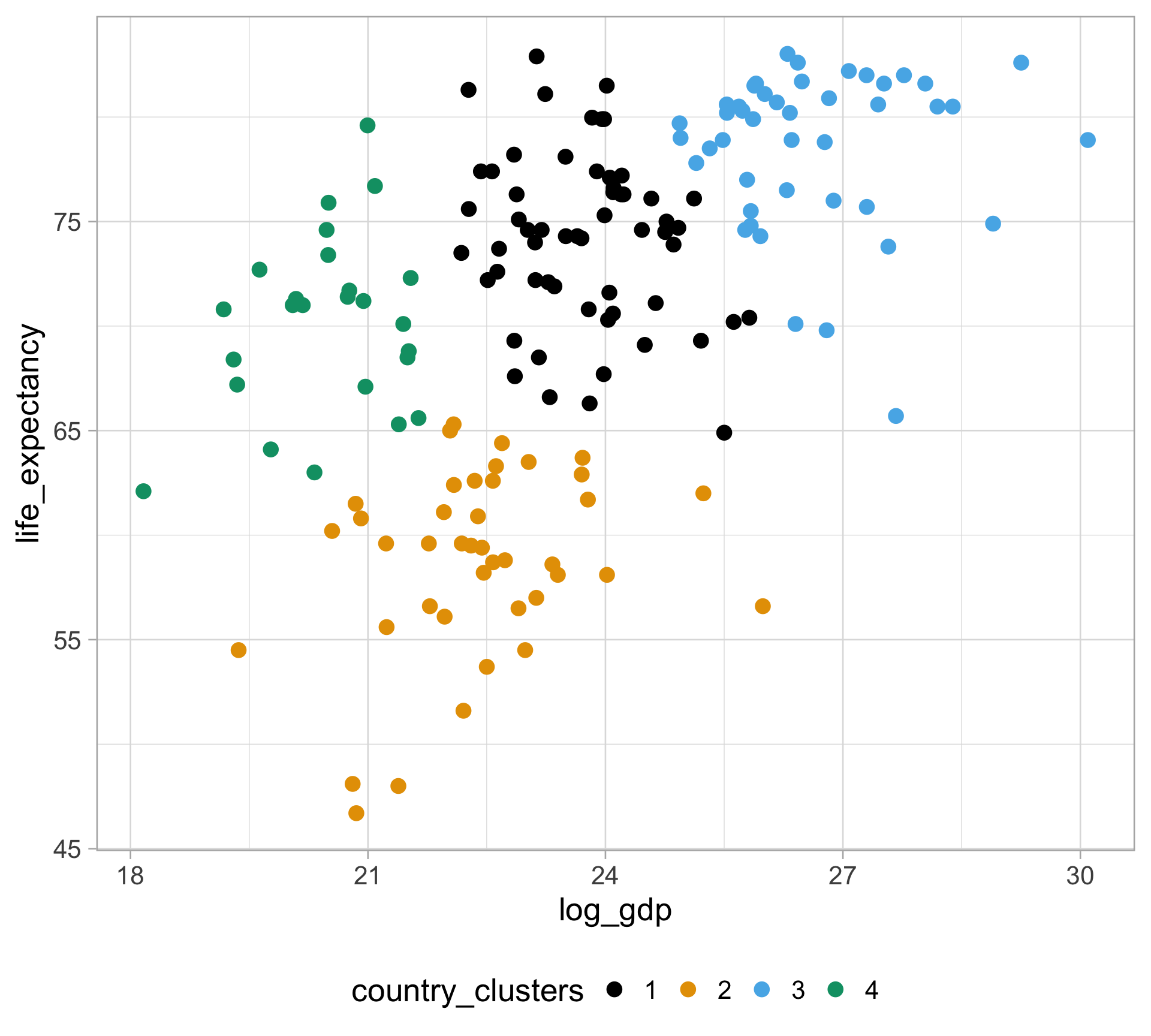
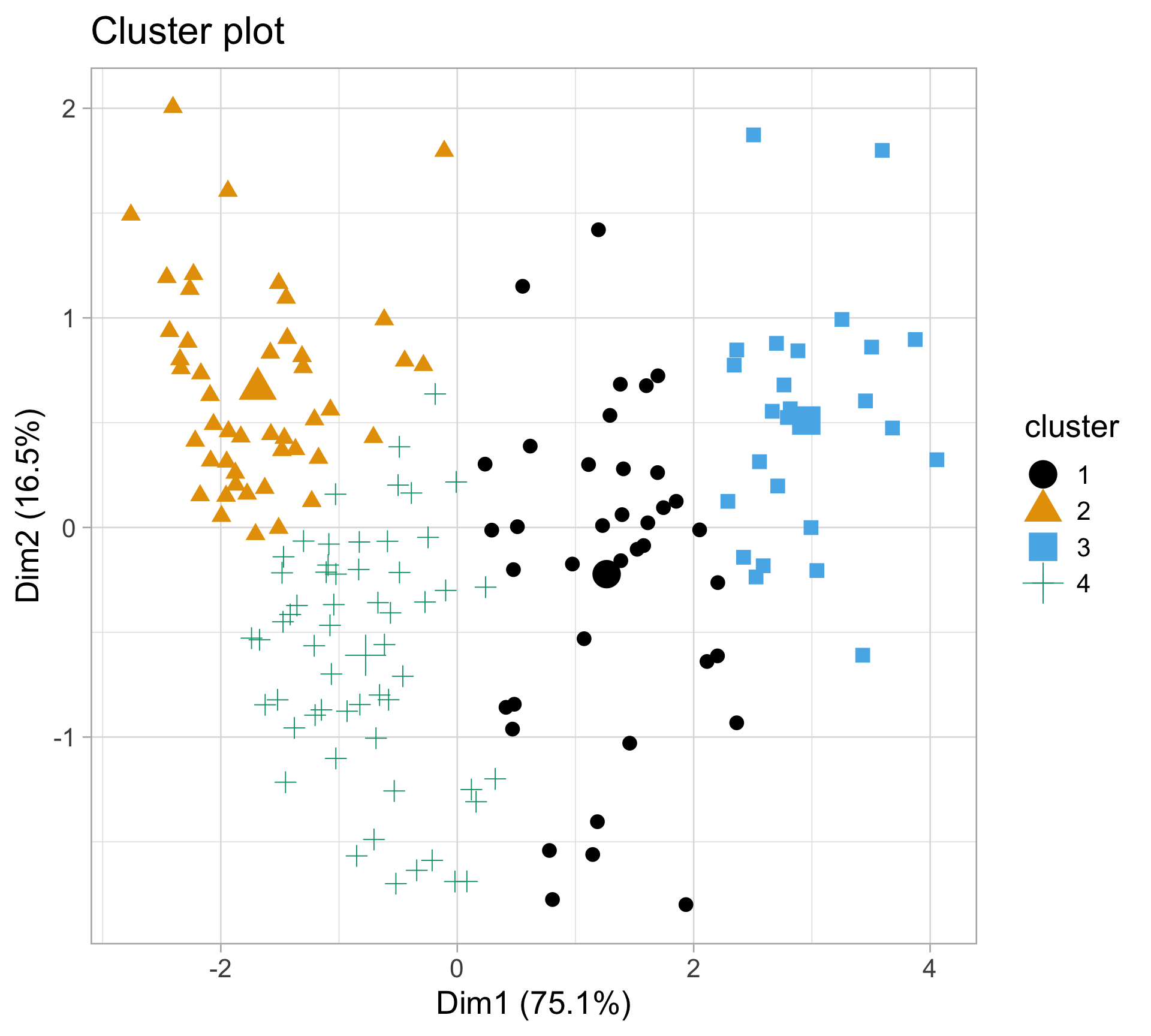
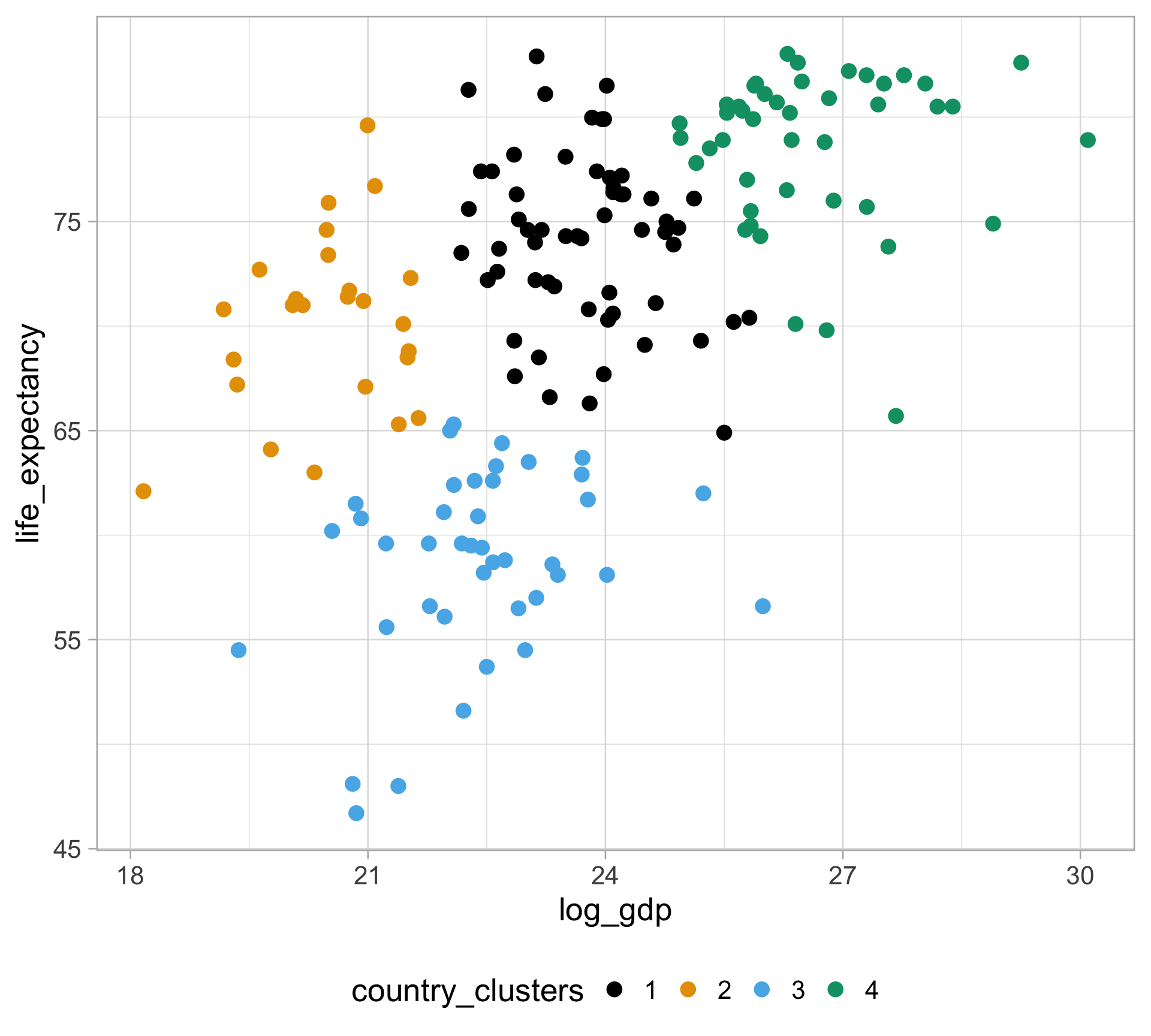
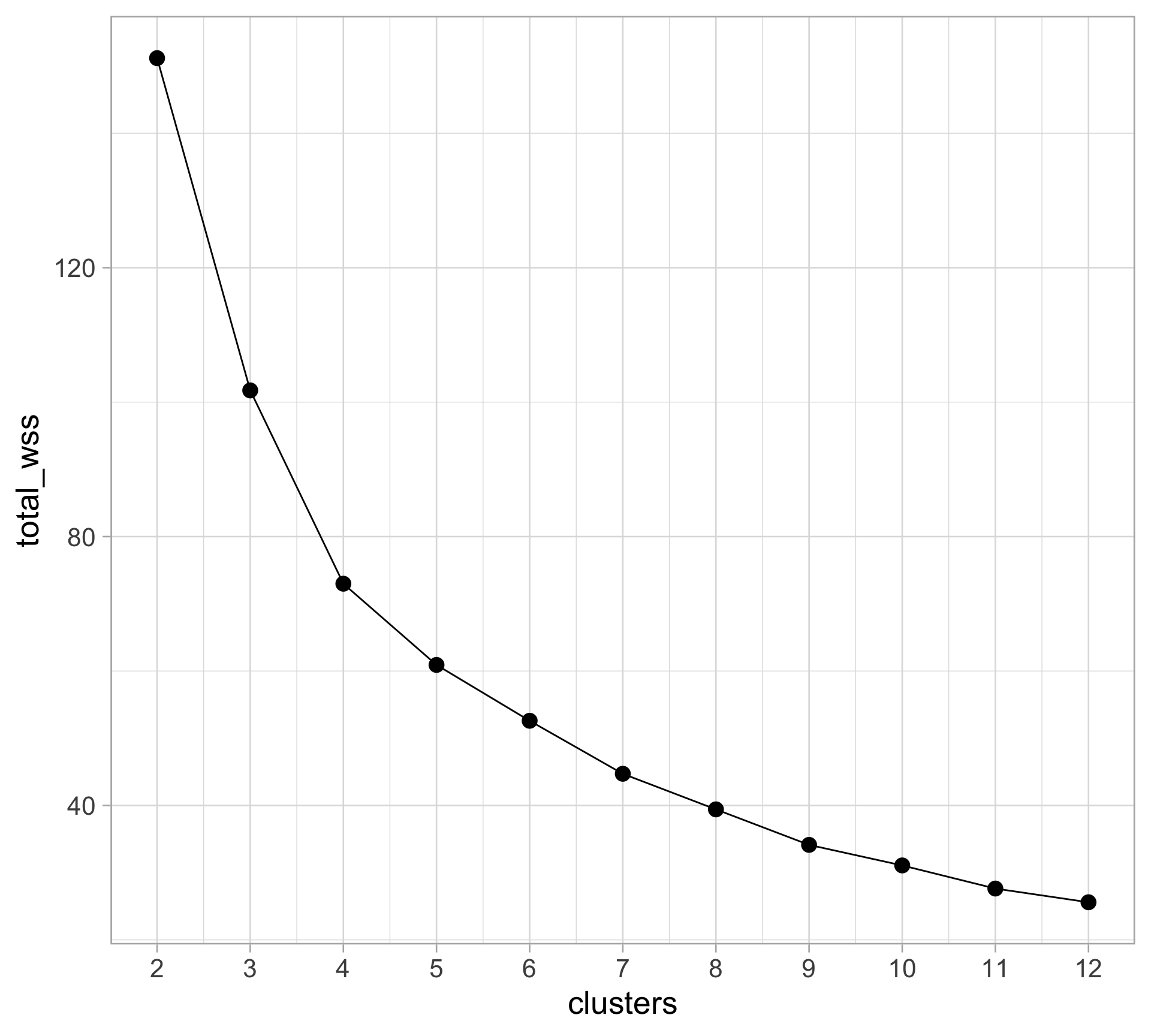
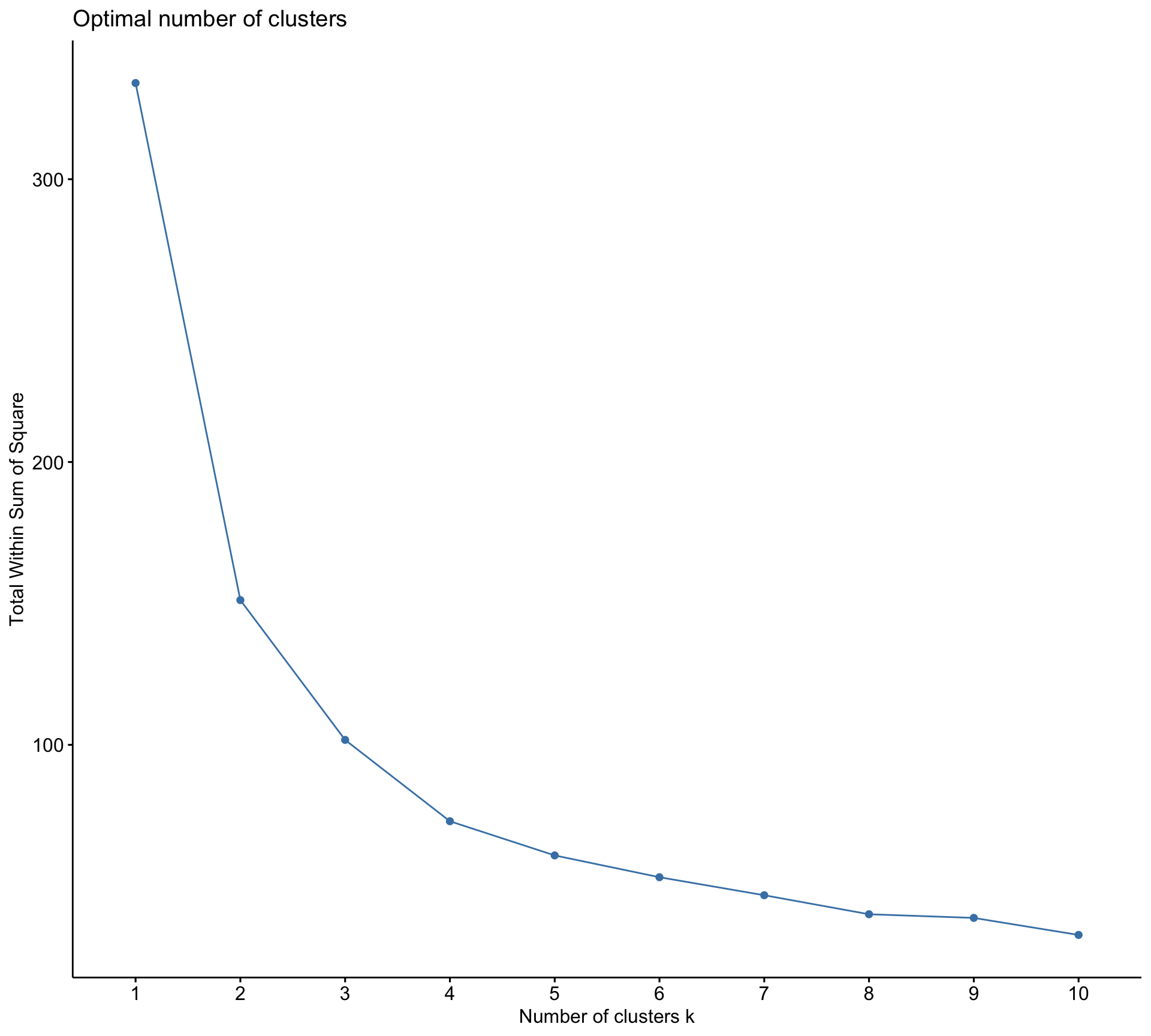
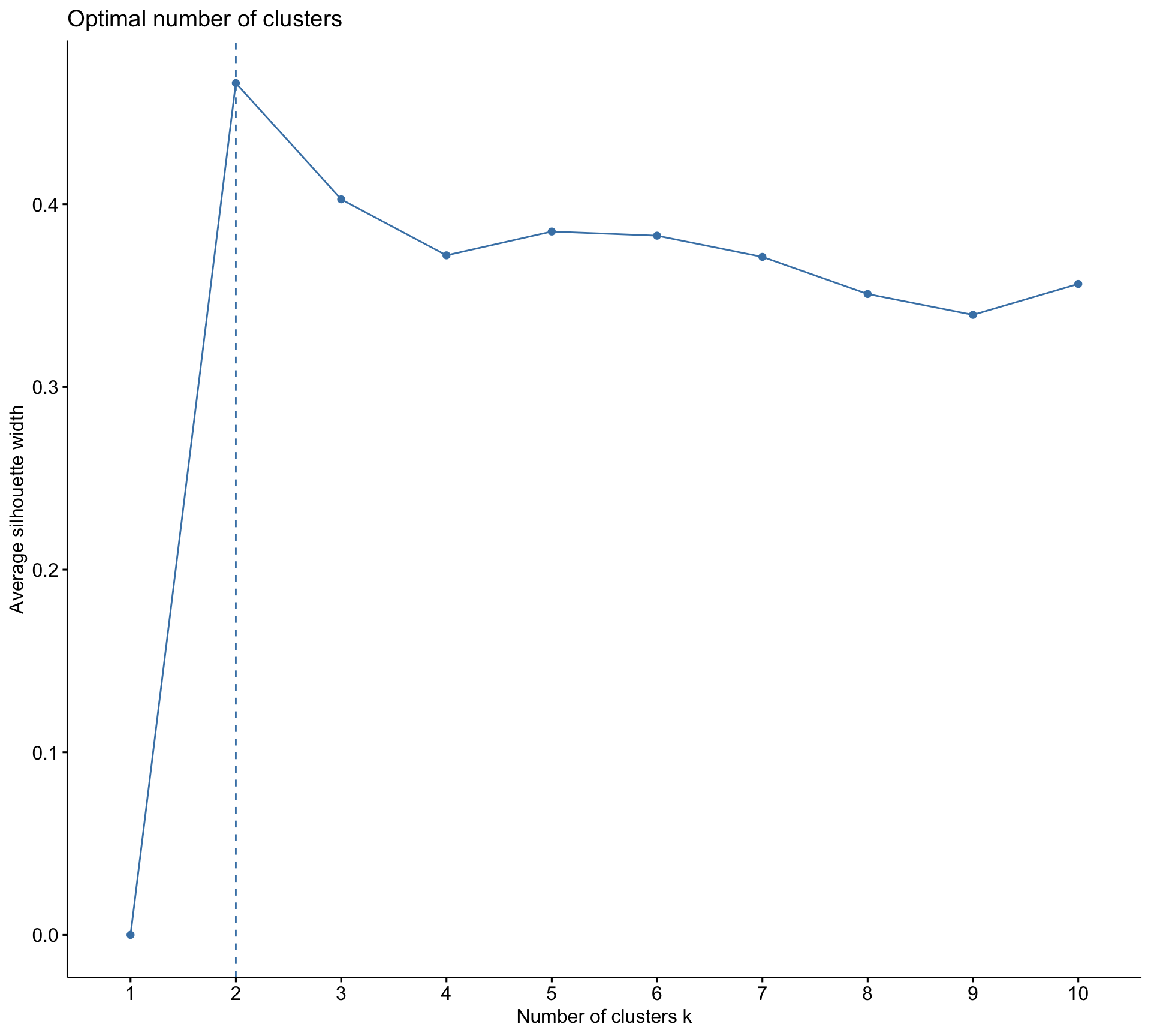
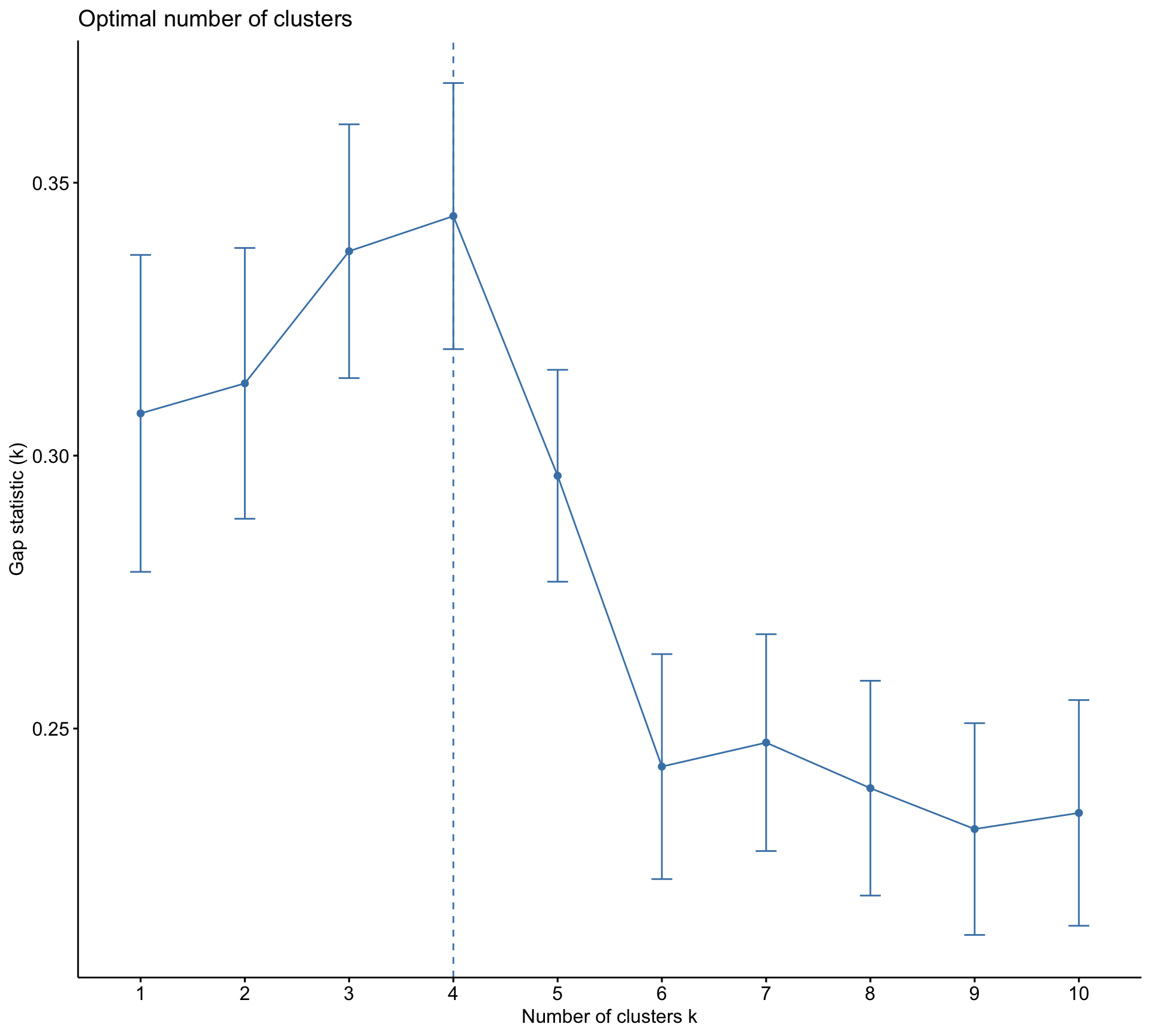
Rows: 10,545
Columns: 9
$ country <fct> "Albania", "Algeria", "Angola", "Antigua and Barbuda"…
$ year <int> 1960, 1960, 1960, 1960, 1960, 1960, 1960, 1960, 1960,…
$ infant_mortality <dbl> 115.40, 148.20, 208.00, NA, 59.87, NA, NA, 20.30, 37.…
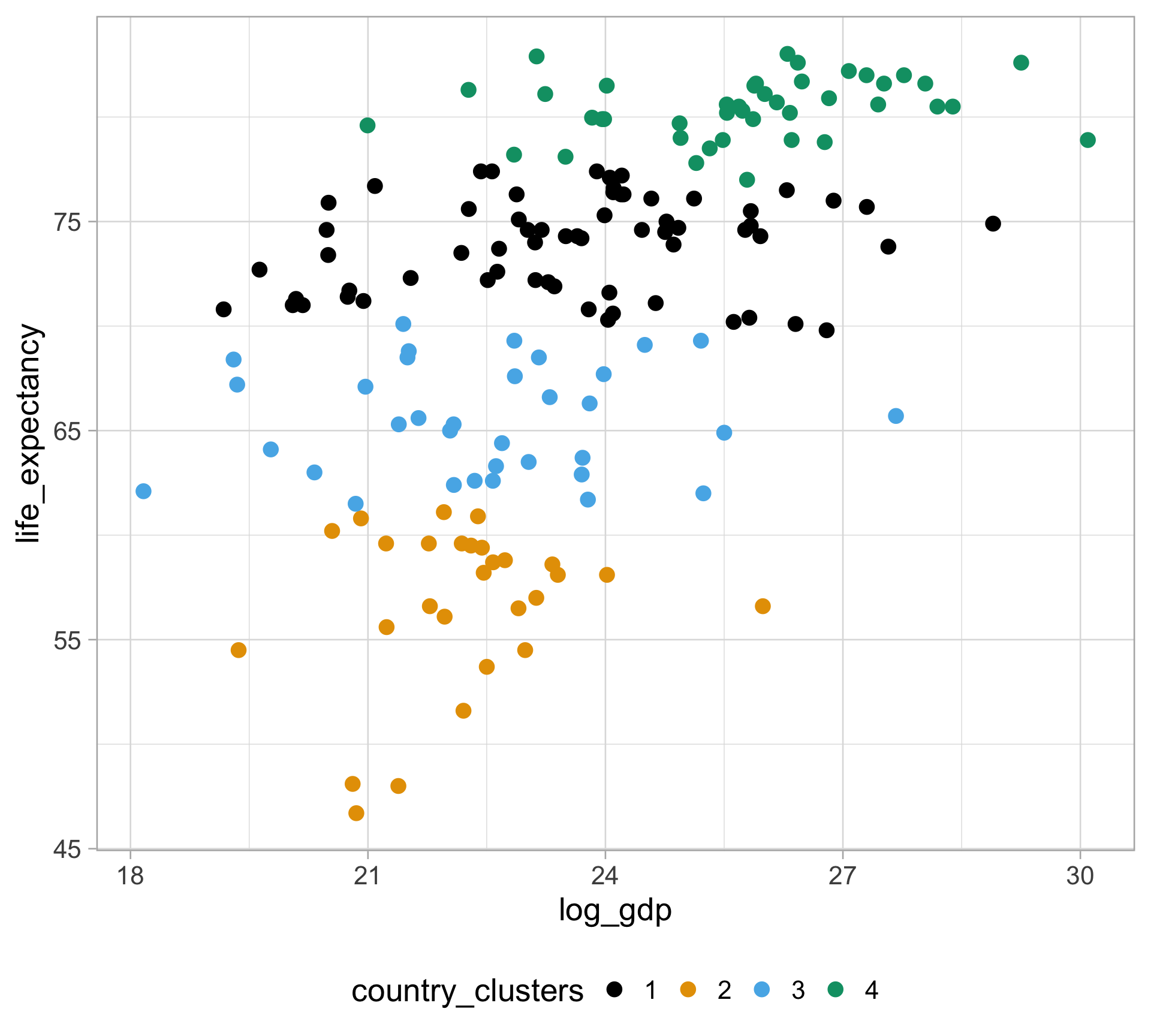
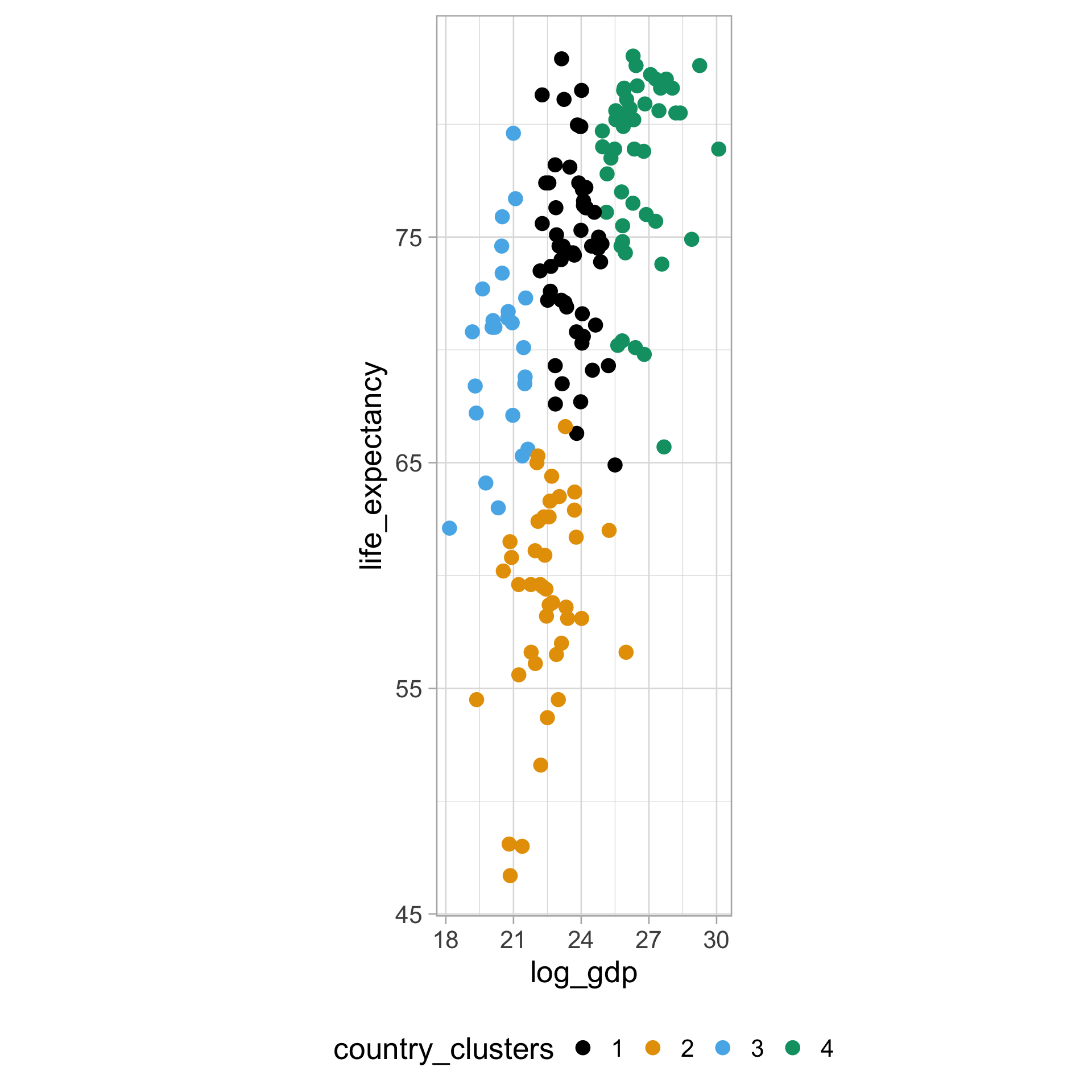
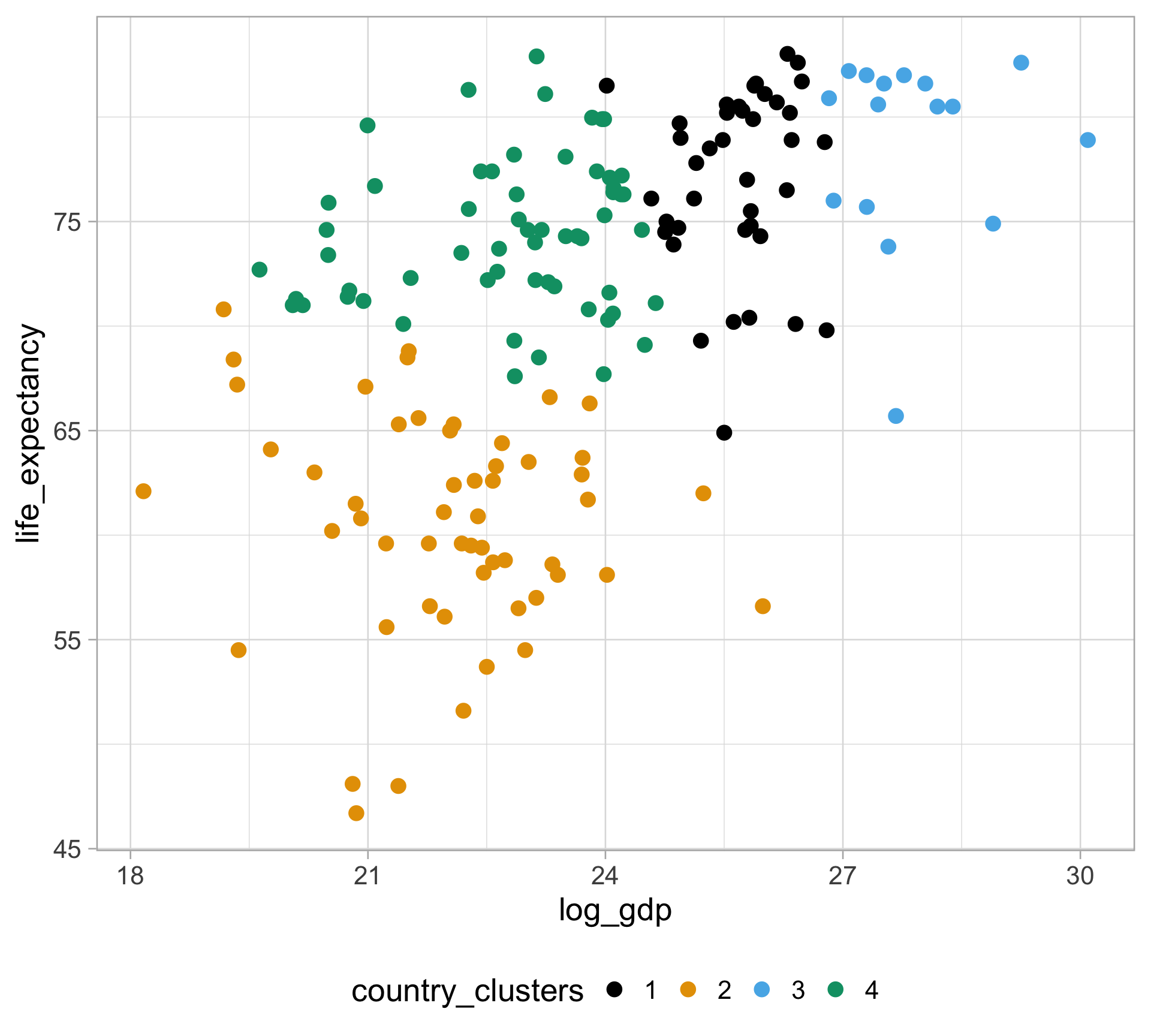
$ life_expectancy <dbl> 62.87, 47.50, 35.98, 62.97, 65.39, 66.86, 65.66, 70.8…
$ fertility <dbl> 6.19, 7.65, 7.32, 4.43, 3.11, 4.55, 4.82, 3.45, 2.70,…
$ population <dbl> 1636054, 11124892, 5270844, 54681, 20619075, 1867396,…
$ gdp <dbl> NA, 13828152297, NA, NA, 108322326649, NA, NA, 966778…
$ continent <fct> Europe, Africa, Africa, Americas, Americas, Asia, Ame…
$ region <fct> Southern Europe, Northern Africa, Middle Africa, Cari…