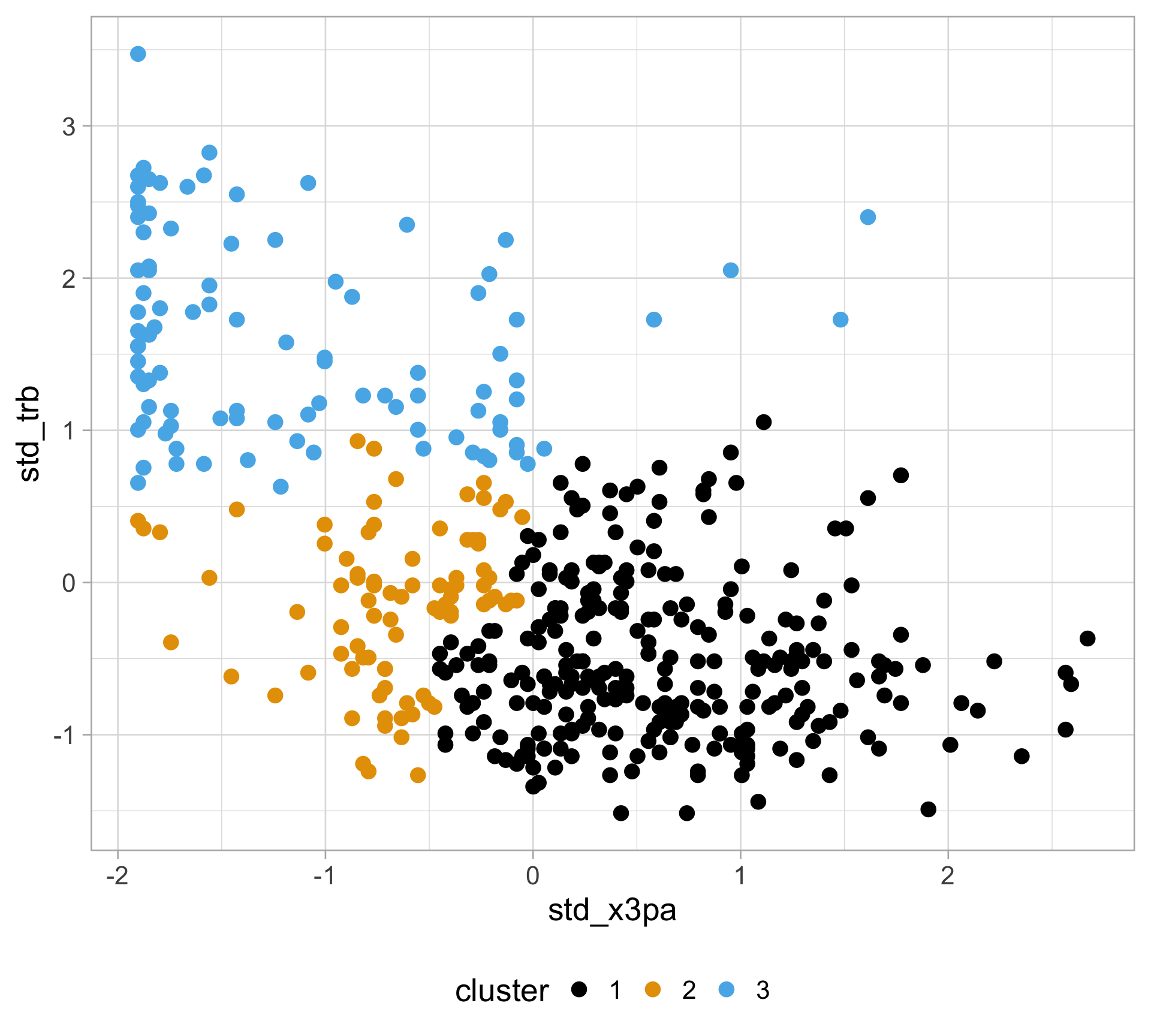
Rows: 457
Columns: 33
$ rk <dbl> 152, 379, 338, 185, 305, 250, 139, 457, 287, 349, 153, 29…
$ player <chr> "A.J. Green", "A.J. Lawson", "AJ Johnson", "Aaron Gordon"…
$ age <dbl> 25, 24, 20, 29, 28, 25, 26, 25, 21, 22, 38, 33, 30, 31, 2…
$ team <chr> "MIL", "TOR", "2TM", "DEN", "HOU", "IND", "OKC", "OKC", "…
$ pos <chr> "SG", "SG", "SG", "PF", "PG", "SF", "SG", "SG", "C", "SG"…
$ g <dbl> 73, 26, 29, 51, 62, 45, 76, 37, 58, 36, 60, 49, 54, 46, 1…
$ gs <dbl> 7, 2, 11, 42, 3, 37, 26, 0, 11, 1, 42, 14, 3, 7, 0, 67, 7…
$ mp <dbl> 1659, 486, 639, 1447, 792, 1123, 1744, 203, 905, 597, 165…
$ fg <dbl> 5.3, 7.9, 6.1, 8.8, 7.2, 8.2, 9.8, 5.9, 7.4, 7.3, 5.9, 6.…
$ fga <dbl> 12.4, 18.8, 15.9, 16.5, 16.5, 16.2, 20.0, 22.7, 10.5, 14.…
$ fg_percent <dbl> 0.429, 0.421, 0.385, 0.531, 0.437, 0.507, 0.488, 0.260, 0…
$ x3p <dbl> 4.5, 3.3, 1.8, 2.5, 4.4, 3.6, 3.6, 3.3, 0.0, 1.8, 3.4, 5.…
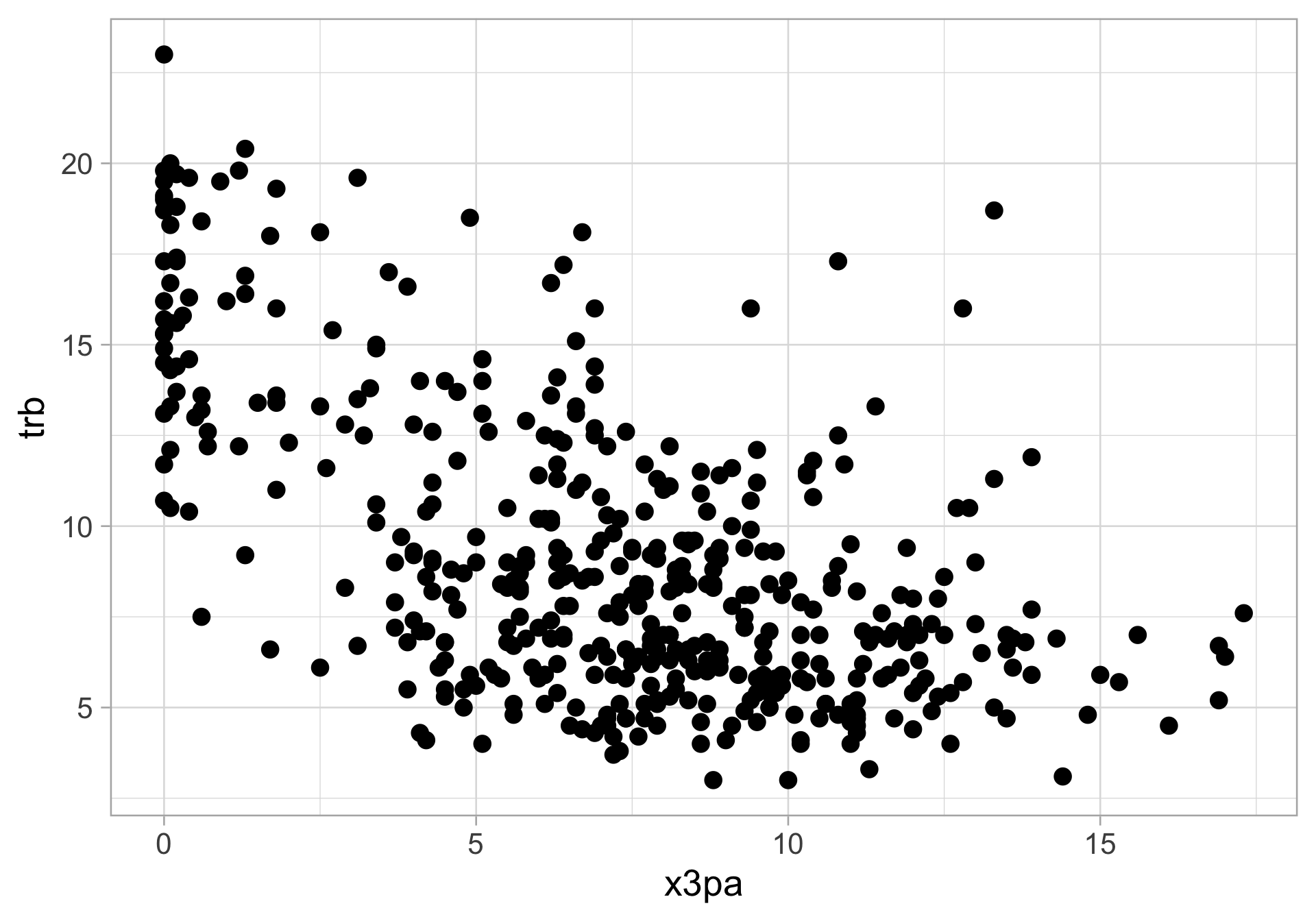
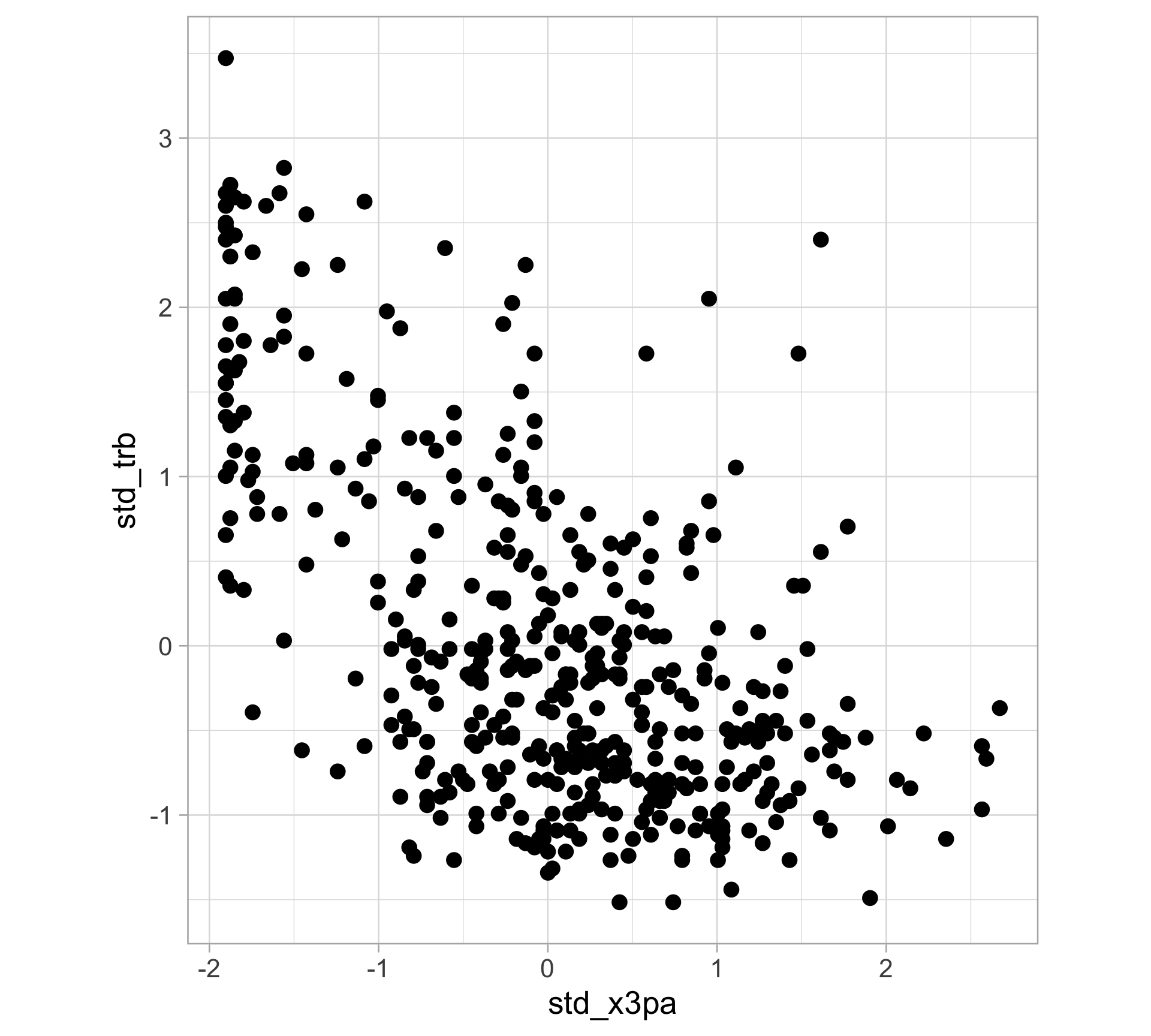
$ x3pa <dbl> 10.6, 10.0, 6.7, 5.7, 11.1, 8.3, 9.3, 17.0, 0.1, 4.8, 9.5…
$ x3p_percent <dbl> 0.427, 0.327, 0.267, 0.436, 0.398, 0.431, 0.383, 0.194, 0…
$ x2p <dbl> 0.8, 4.7, 4.3, 6.3, 2.8, 4.6, 6.2, 2.6, 7.4, 5.5, 2.4, 1.…
$ x2pa <dbl> 1.8, 8.8, 9.2, 10.8, 5.3, 7.9, 10.7, 5.7, 10.4, 10.0, 4.4…
$ x2p_percent <dbl> 0.443, 0.528, 0.472, 0.582, 0.517, 0.587, 0.578, 0.458, 0…
$ e_fg_percent <dbl> 0.612, 0.508, 0.441, 0.607, 0.571, 0.617, 0.577, 0.333, 0…
$ ft <dbl> 0.6, 4.3, 2.4, 4.8, 2.1, 3.1, 2.0, 0.2, 3.6, 2.3, 1.0, 2.…
$ fta <dbl> 0.8, 6.2, 2.8, 5.9, 2.5, 3.4, 2.4, 0.5, 5.4, 2.8, 1.1, 2.…
$ ft_percent <dbl> 0.815, 0.683, 0.865, 0.810, 0.829, 0.913, 0.831, 0.500, 0…
$ orb <dbl> 0.5, 2.0, 0.6, 2.7, 0.8, 1.6, 2.2, 1.7, 5.0, 1.4, 2.4, 0.…
$ drb <dbl> 4.5, 6.5, 3.8, 5.5, 4.0, 6.0, 5.9, 4.7, 8.3, 4.0, 8.8, 6.…
$ trb <dbl> 5.1, 8.5, 4.4, 8.2, 4.8, 7.6, 8.1, 6.4, 13.3, 5.5, 11.2, …
$ ast <dbl> 3.1, 3.1, 5.7, 5.5, 5.1, 2.3, 3.7, 2.8, 1.5, 5.1, 3.9, 3.…
$ stl <dbl> 1.1, 1.3, 0.9, 0.8, 1.2, 1.5, 1.7, 1.9, 1.4, 2.0, 1.1, 1.…
$ blk <dbl> 0.2, 0.6, 0.2, 0.5, 0.7, 0.7, 0.5, 0.7, 3.8, 0.3, 1.5, 0.…
$ tov <dbl> 1.2, 1.5, 2.6, 2.4, 2.3, 1.6, 1.9, 0.9, 3.3, 2.3, 1.4, 1.…
$ pf <dbl> 4.6, 4.4, 3.7, 2.7, 3.9, 4.9, 2.8, 3.5, 6.8, 5.5, 2.4, 2.…
$ pts <dbl> 15.8, 23.4, 16.4, 24.9, 20.9, 23.2, 25.2, 15.4, 18.4, 18.…
$ o_rtg <dbl> 119, 113, 98, 129, 121, 124, 119, 80, 123, 117, 121, 117,…
$ d_rtg <dbl> 117, 116, 123, 120, 114, 116, 110, 110, 115, 111, 111, 11…
$ awards <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…