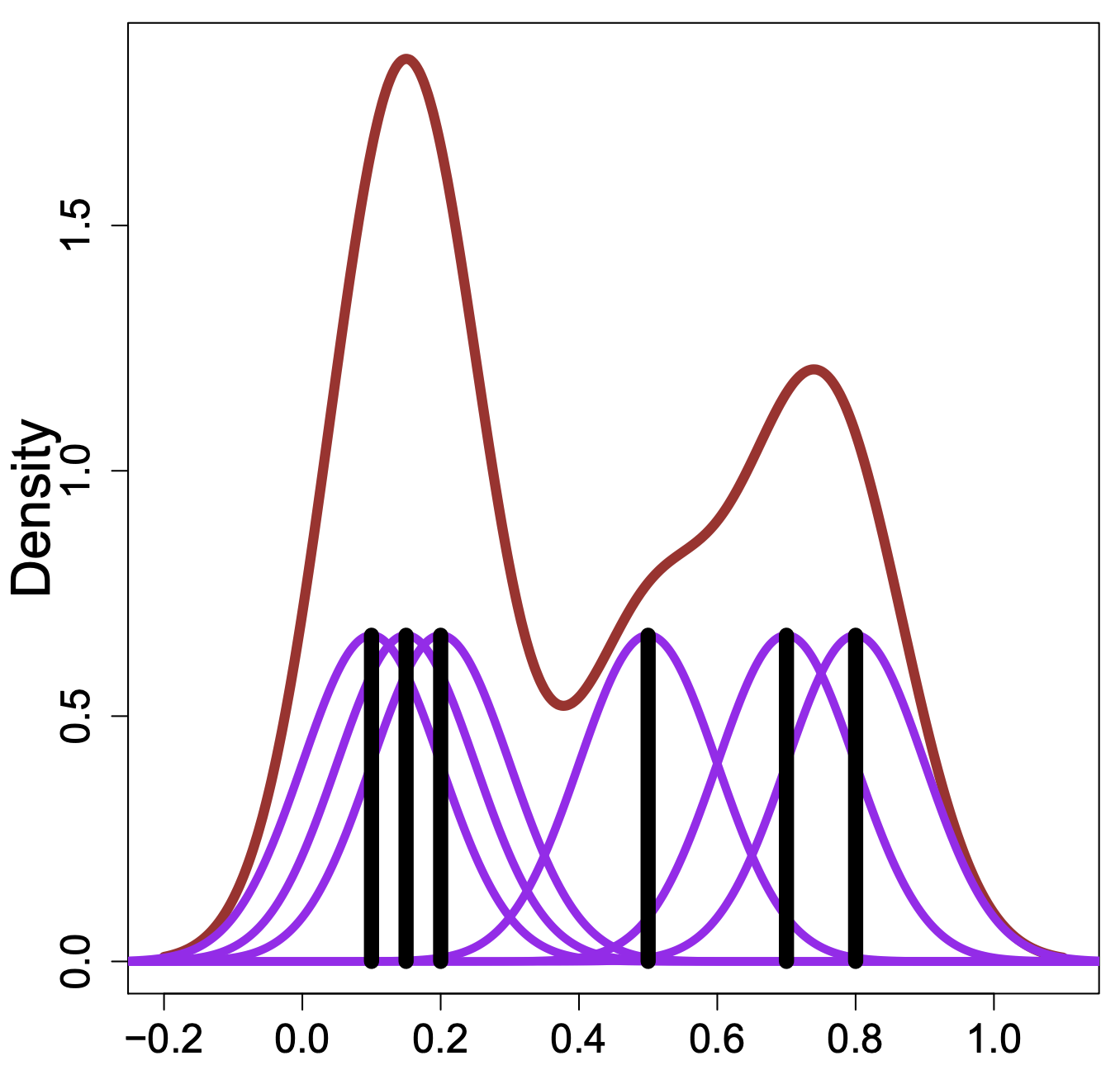
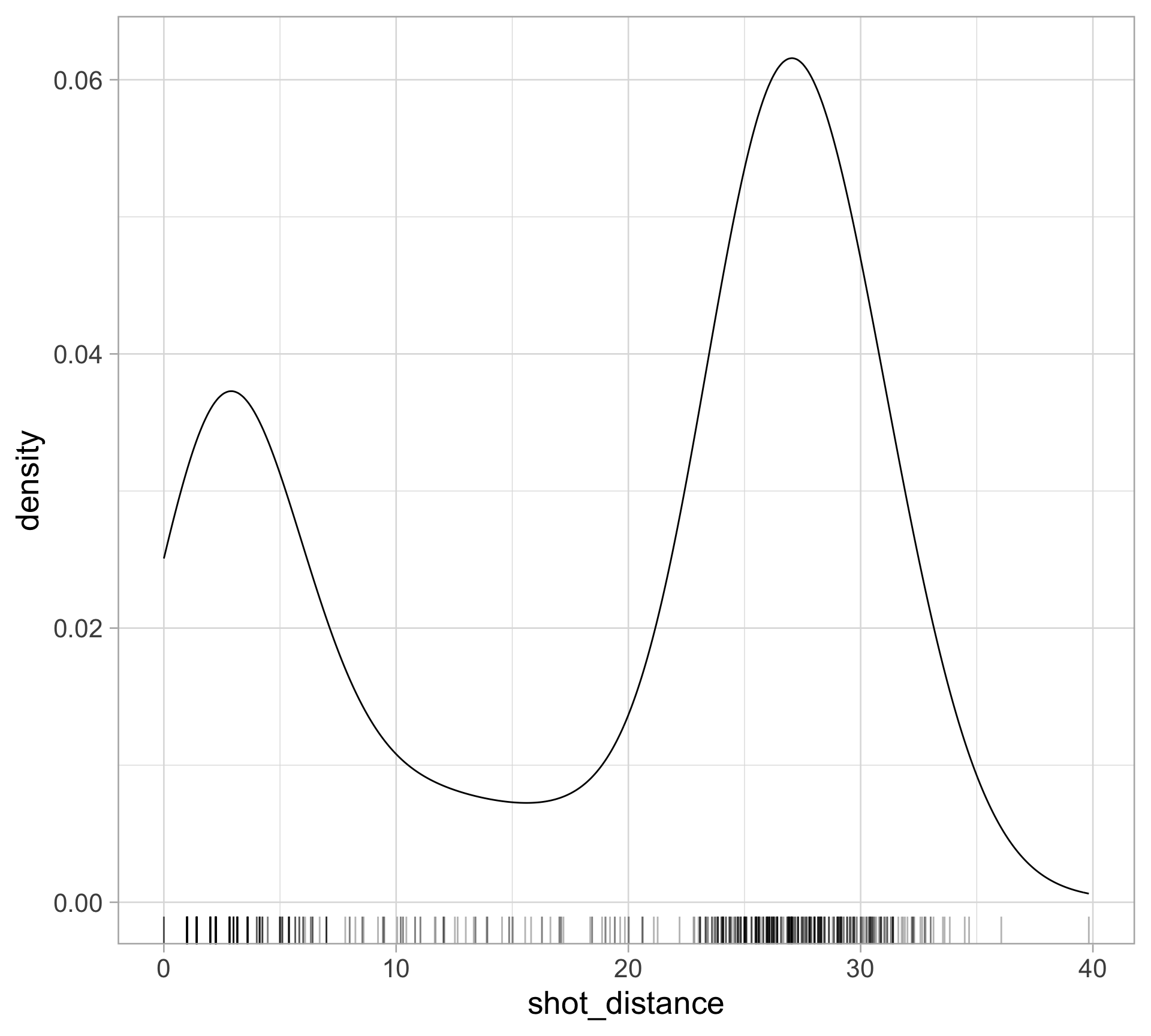
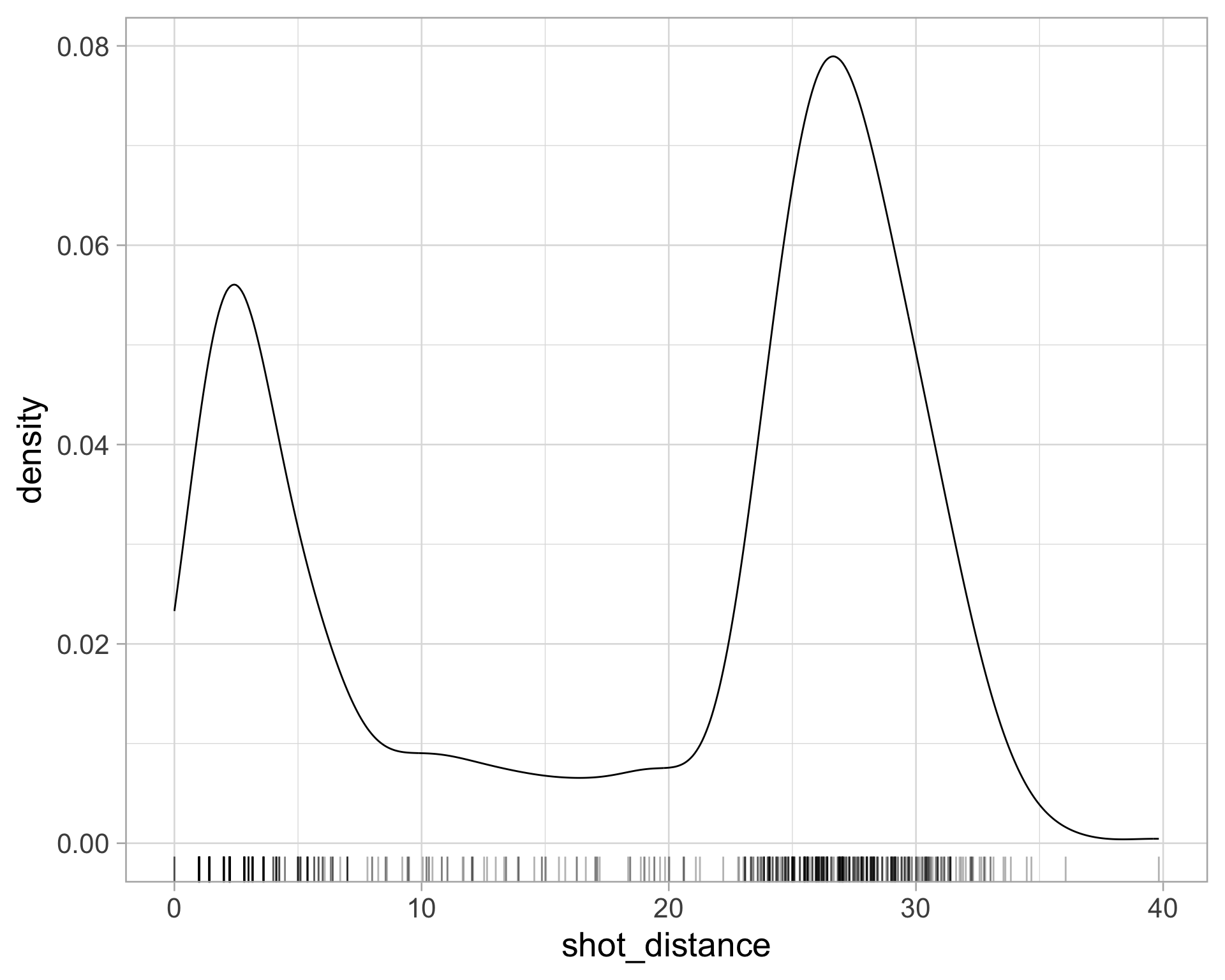
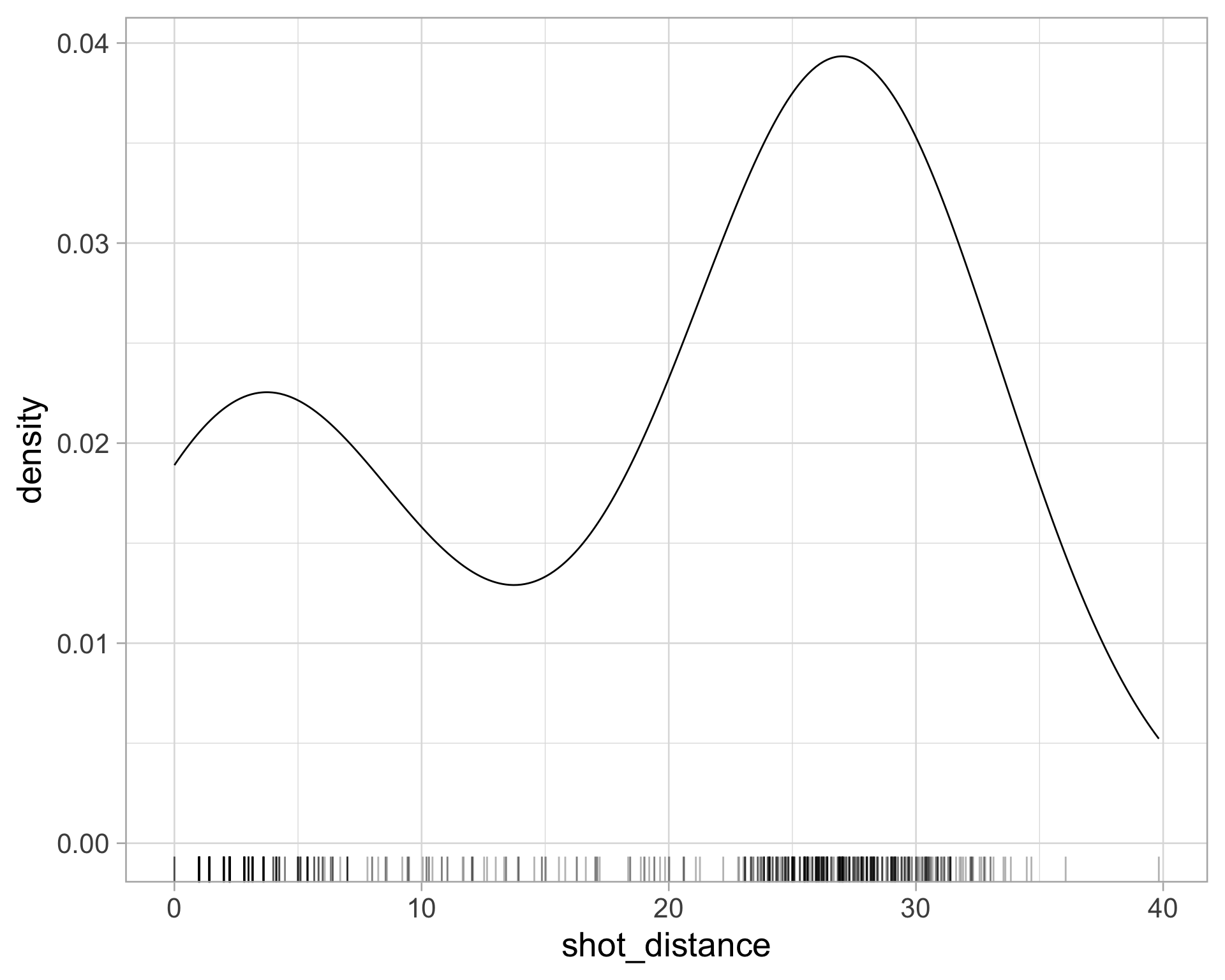
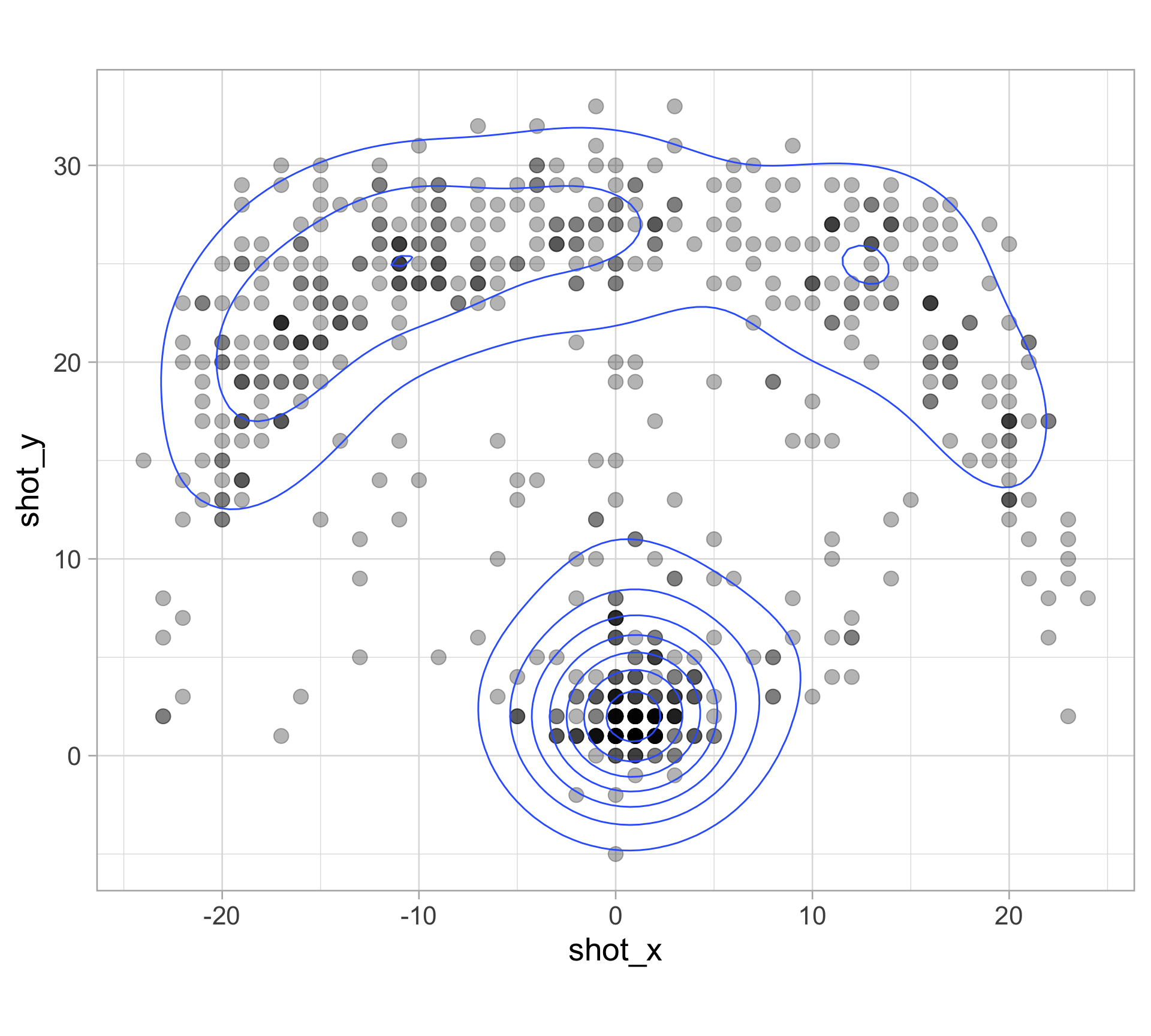
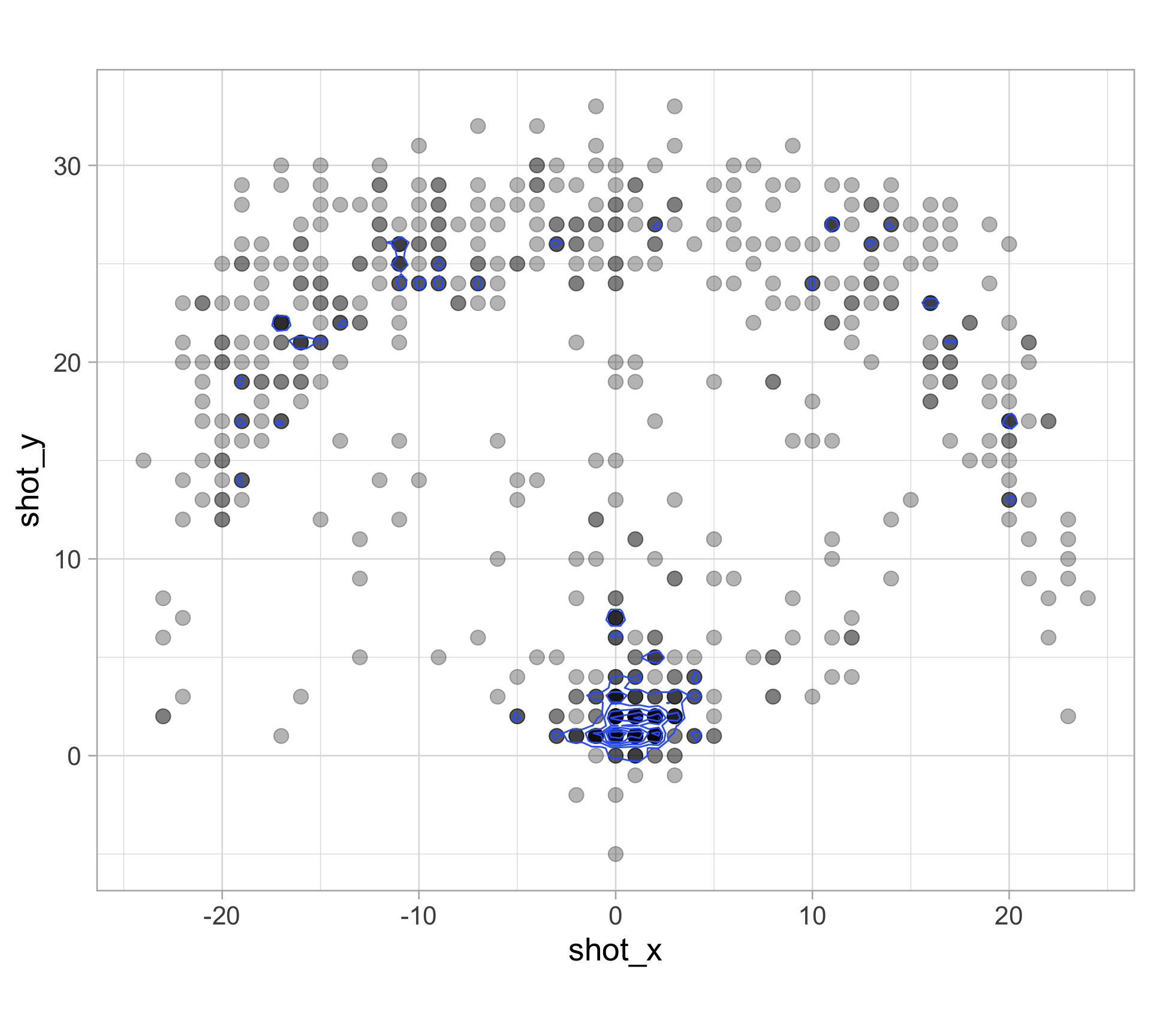
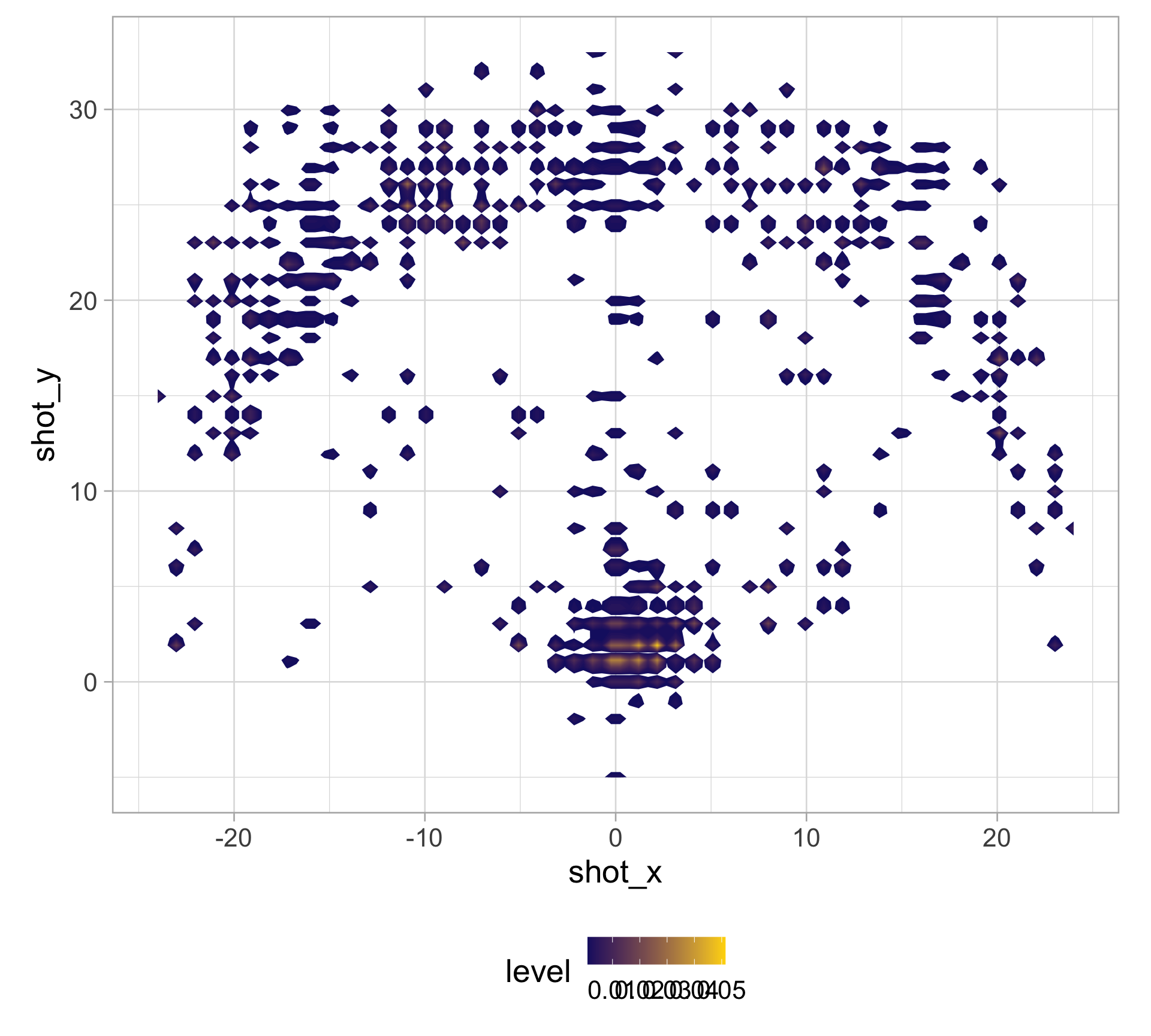
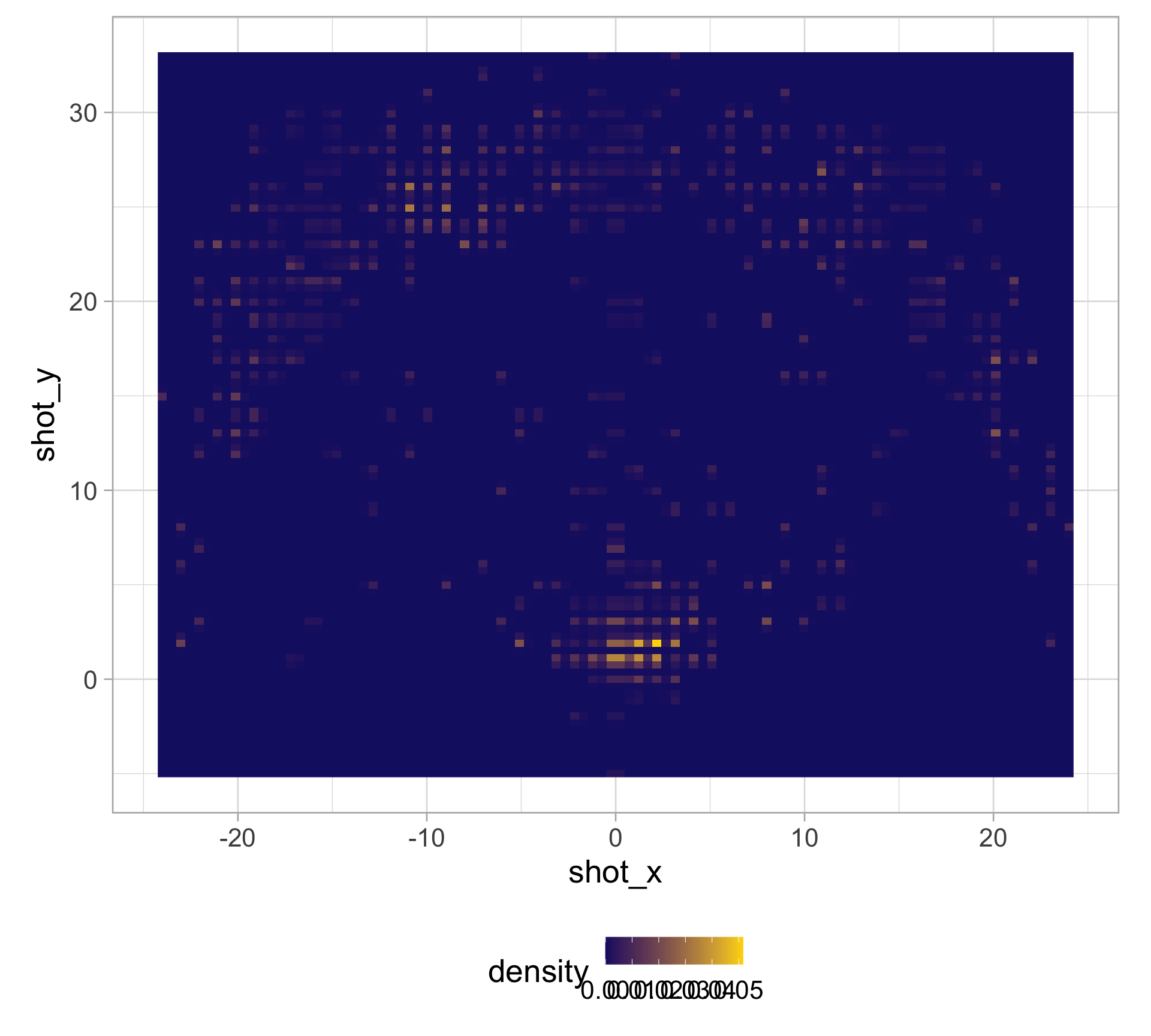
library(tidyverse)
theme_set(theme_light())
clark_shots <- read_csv("https://raw.githubusercontent.com/36-SURE/2025/main/data/clark_shots.csv")
glimpse(clark_shots)Rows: 658
Columns: 6
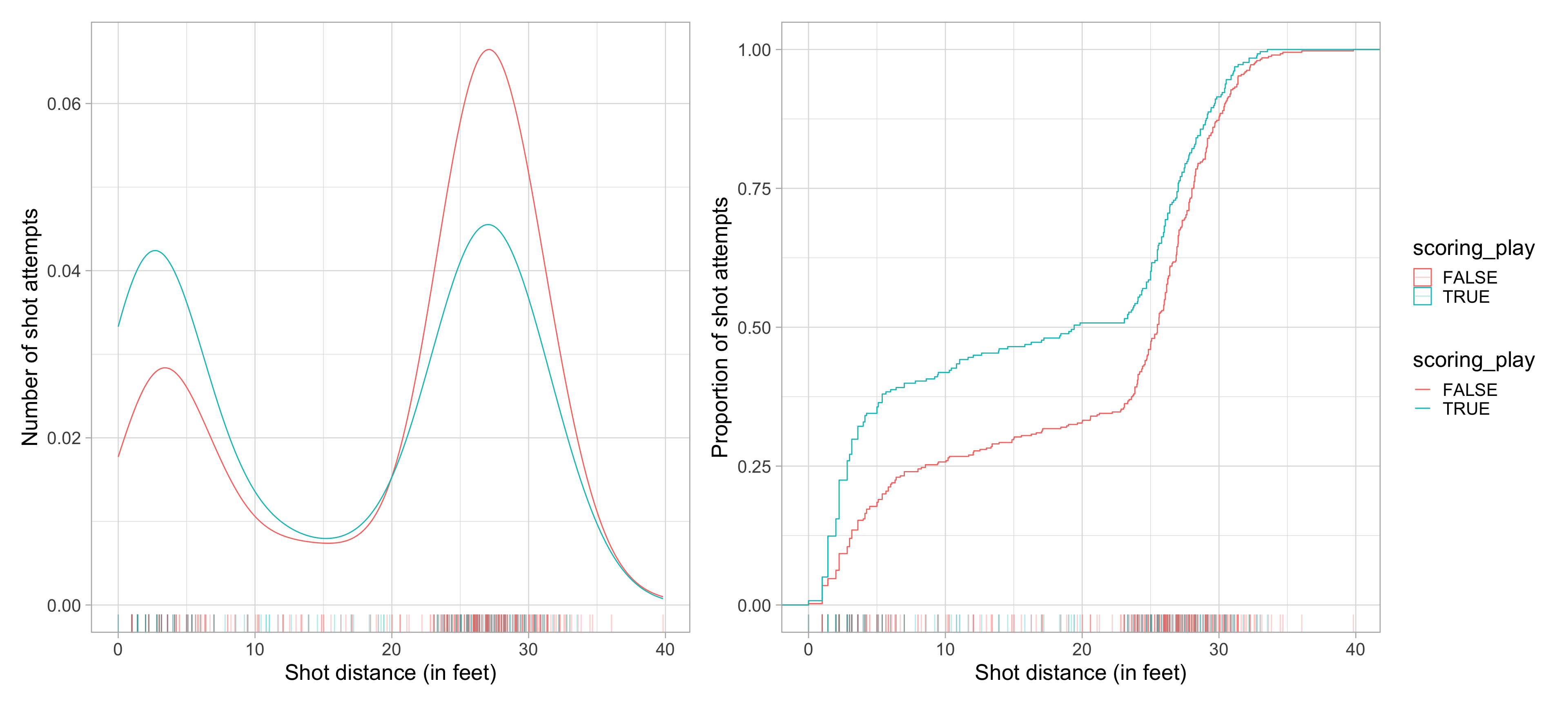
$ scoring_play <lgl> TRUE, FALSE, FALSE, TRUE, FALSE, FALSE, TRUE, FALSE, TRU…
$ score_value <dbl> 3, 0, 0, 3, 0, 0, 2, 0, 2, 0, 0, 2, 2, 0, 2, 0, 2, 0, 0,…
$ shot_x <dbl> -17, -1, -14, -17, -18, -1, 5, 6, 1, -16, -19, 0, 2, -7,…
$ shot_y <dbl> 22, 33, 16, 19, 20, 0, 6, 30, 1, 21, 13, 4, 1, 32, 1, 14…
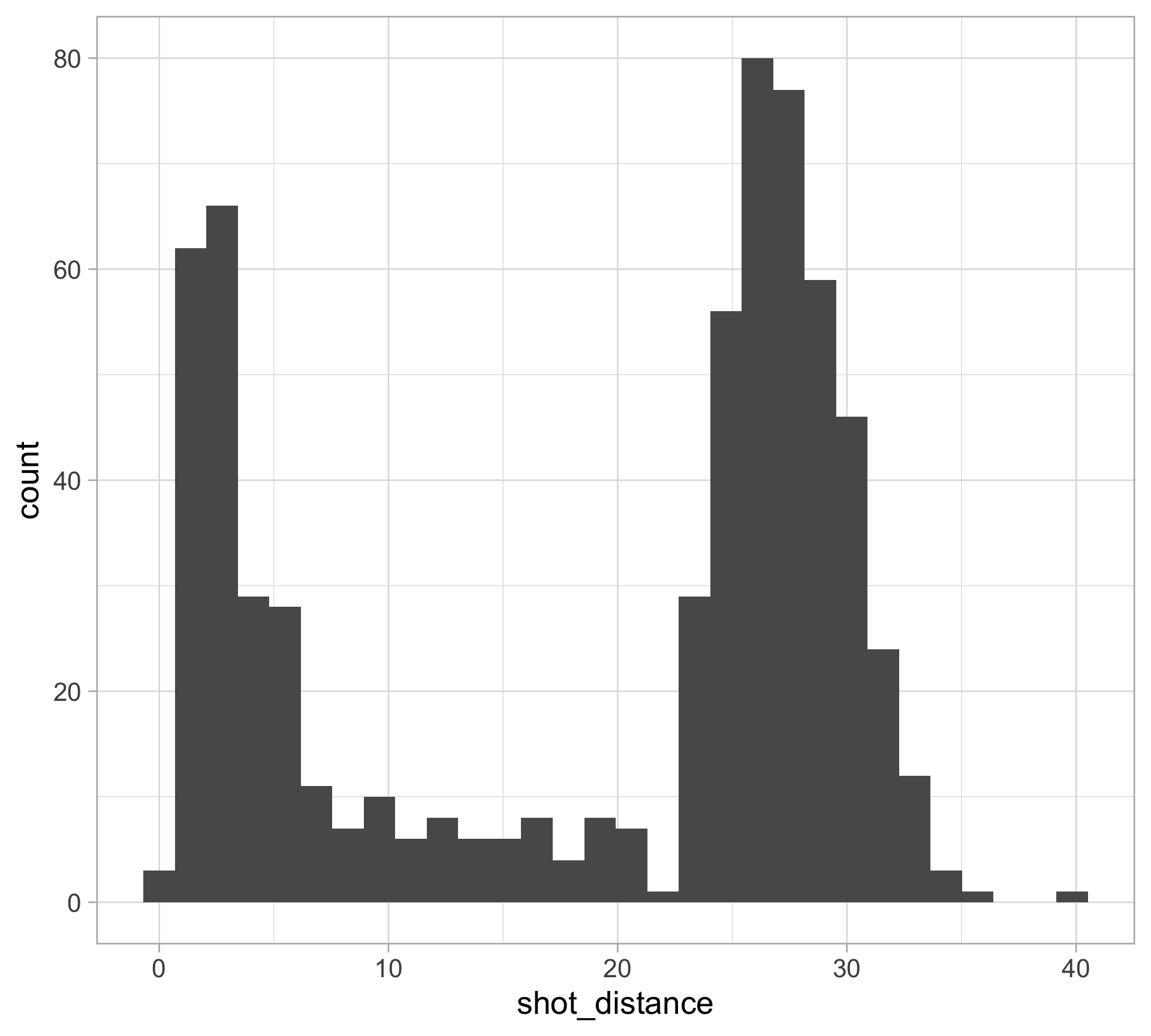
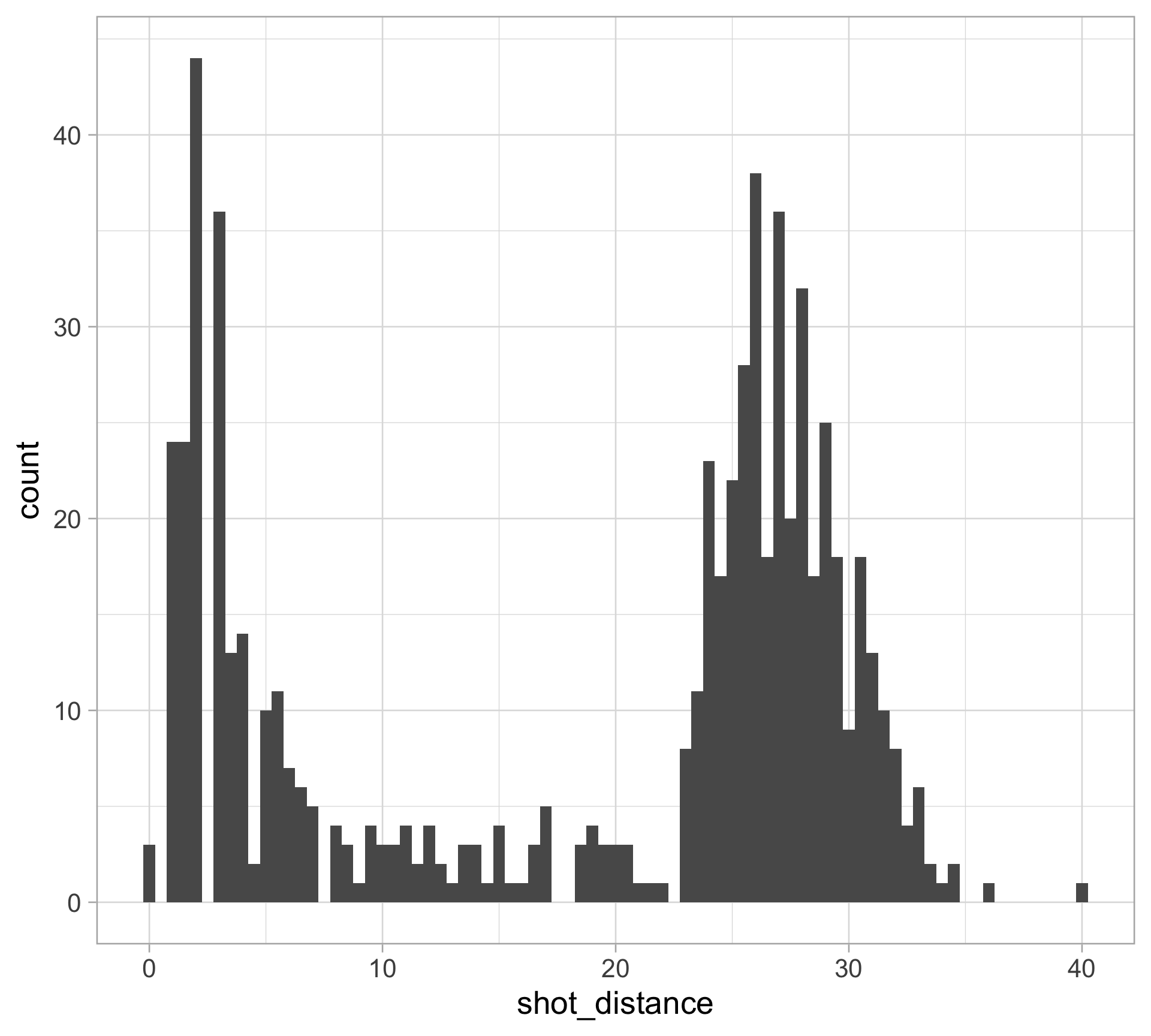
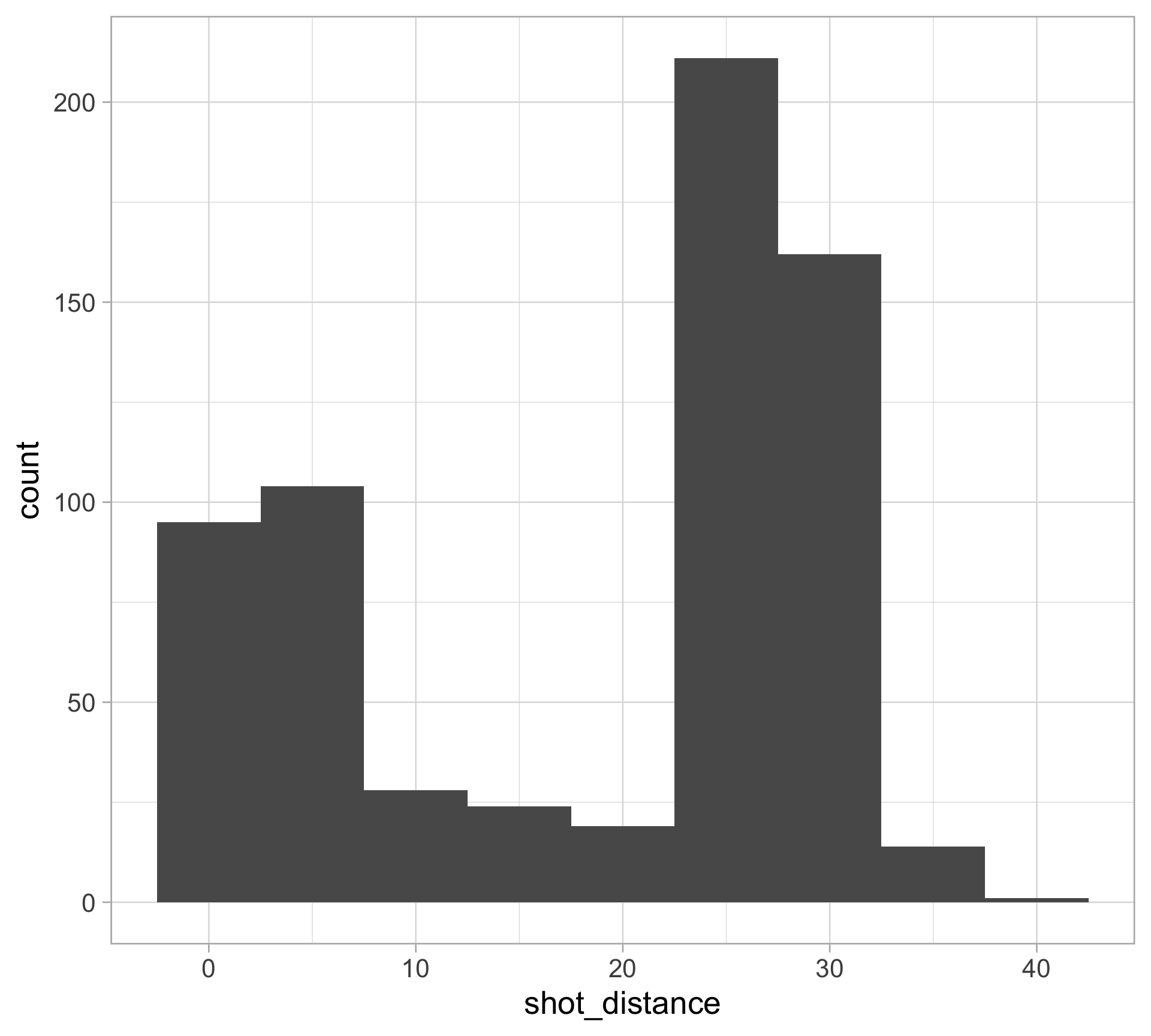
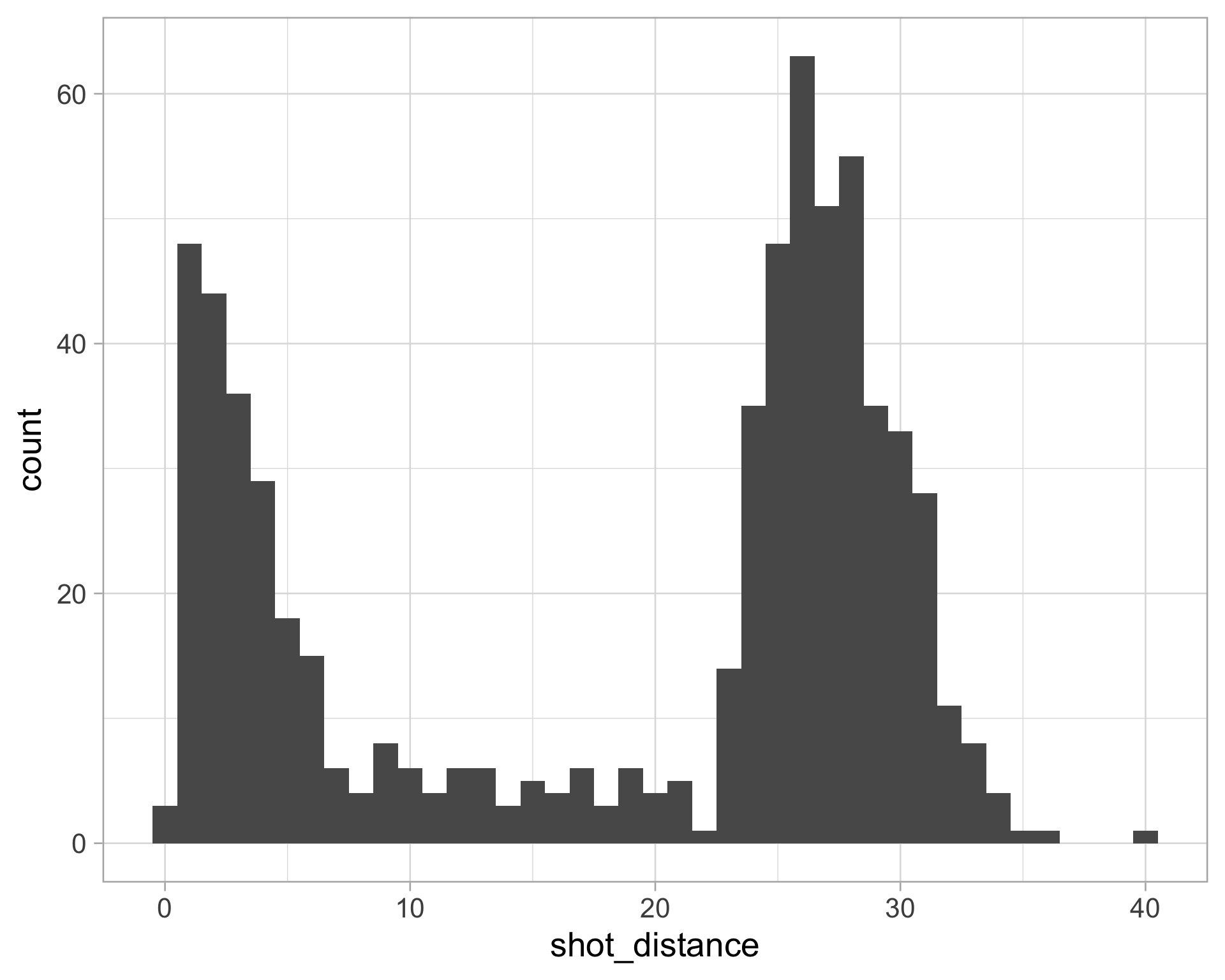
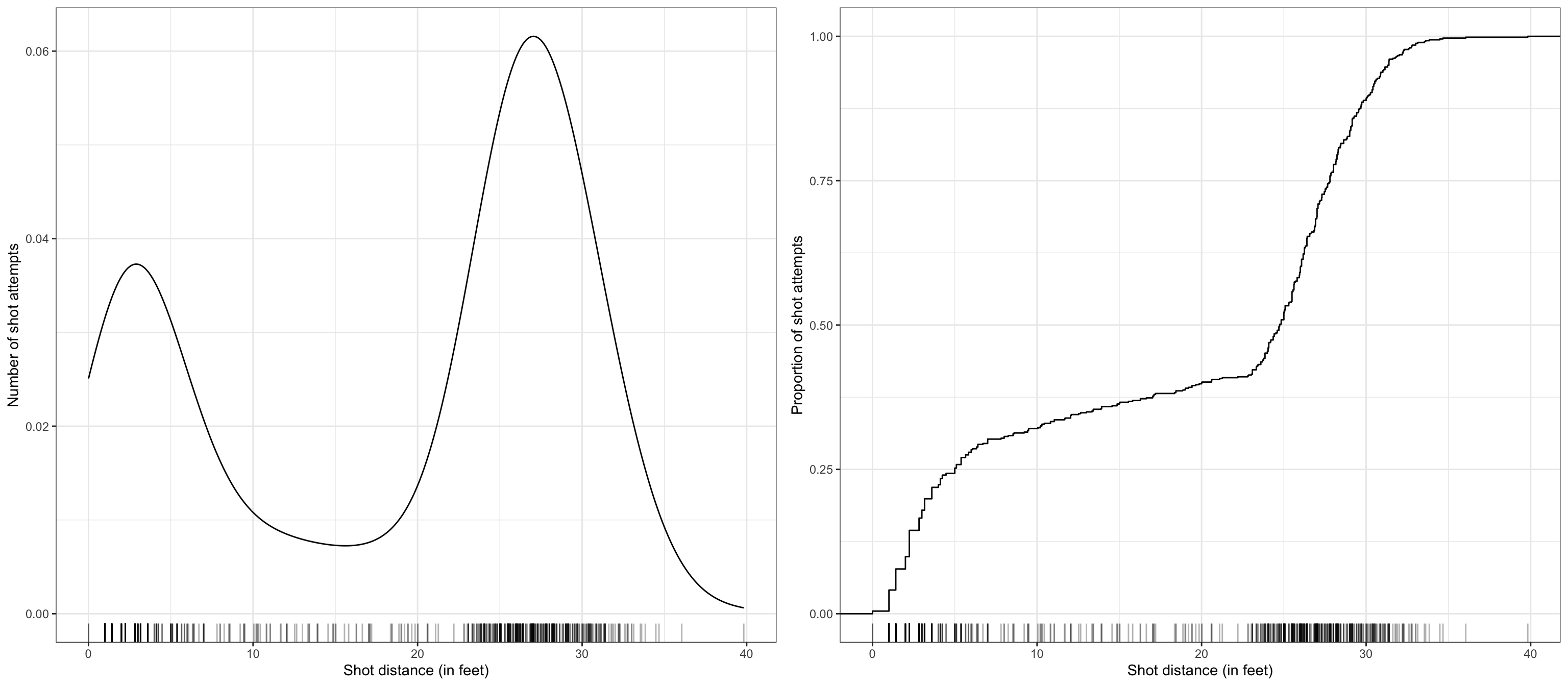
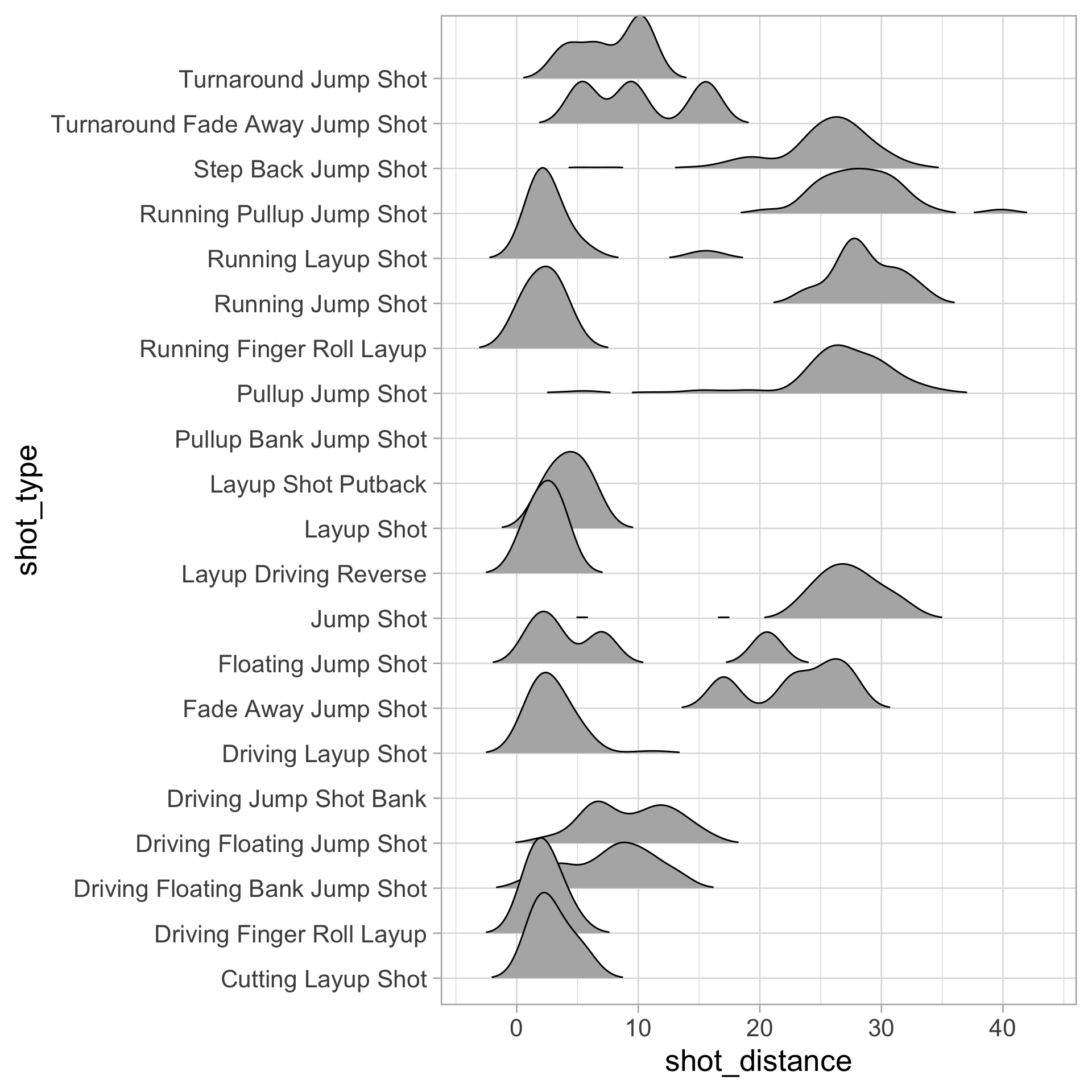
$ shot_distance <dbl> 27.802878, 33.015148, 21.260292, 25.495098, 26.907248, 1…
$ shot_type <chr> "Running Pullup Jump Shot", "Pullup Jump Shot", "Step Ba…