
Data wrangling with R’s data.table package
Sara Colando & Erin Franke
Department of Statistics & Data Science
Carnegie Mellon University
Pre-Lecture Survey
Motivation
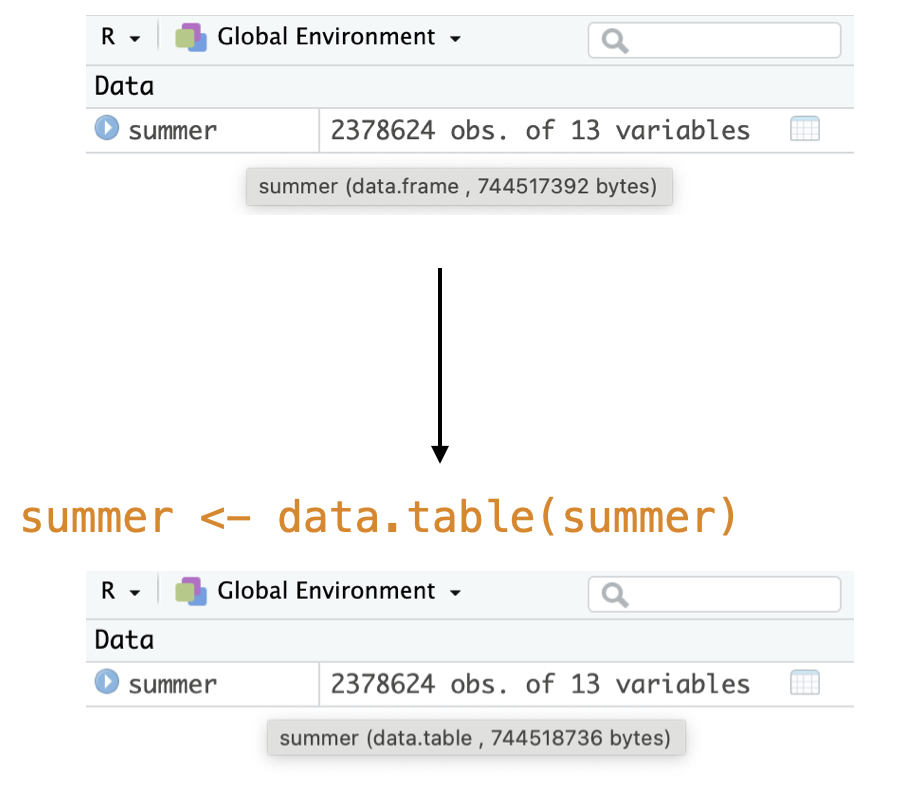
What is data.table?
A versatile R package that is a high performance version of base R’s
data.frame.1Benefits include:
Computational efficiency
Concise syntax
No dependencies
Tested against old versions of R
Uses: data wrangling, reading/writing files, handling large data, and much more!
The six main verbs
Select: extract columns in data
Mutate: create new variables (columns) in data
Filter: extract rows in data that meet certain logical conditions
Arrange: sort rows (observations) by variables
Group by: group the data based on variable(s) in data
Summarize: create summary statistics, usually on grouped data
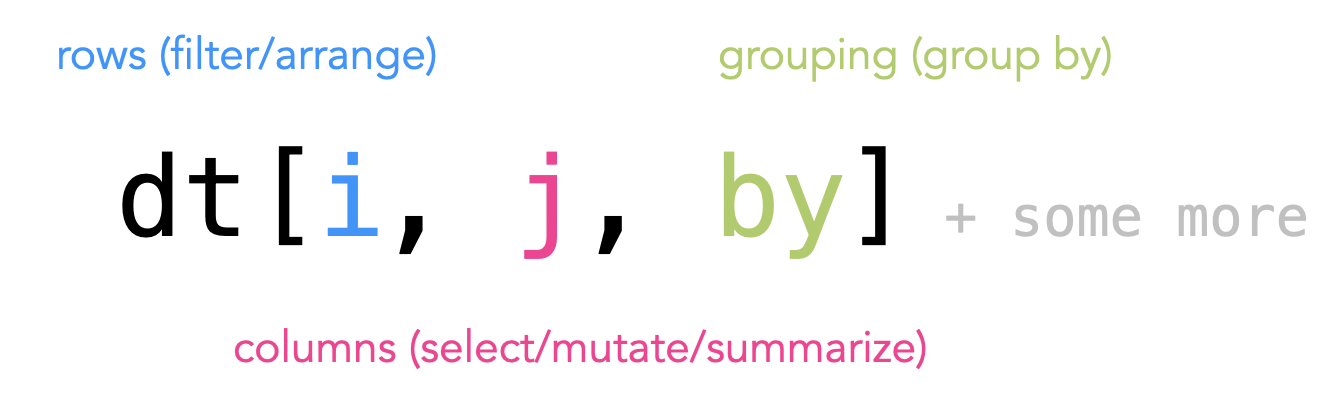
data.table syntax
The six main verbs all follow a three part syntax:
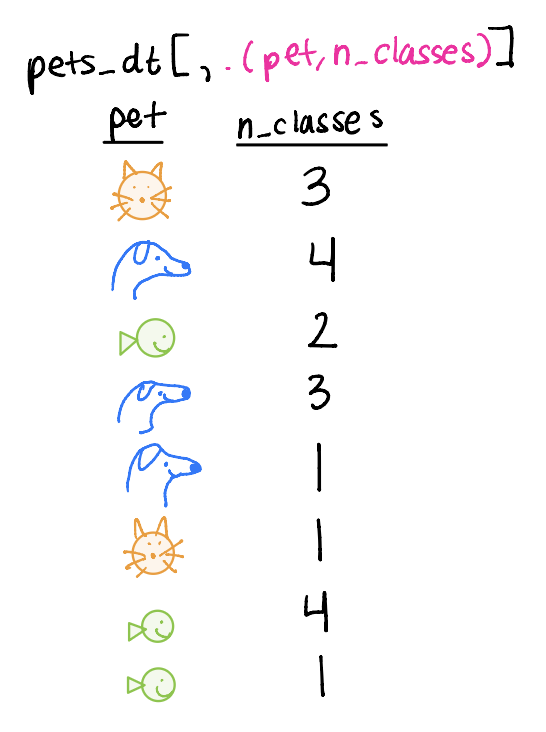
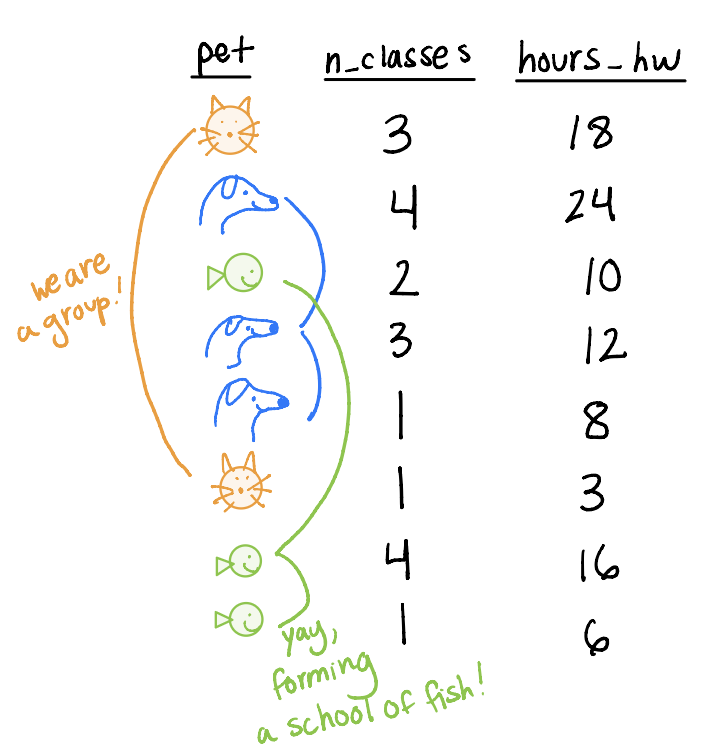
Example data
pets_dtis collected on students who have one petpet: animal student hasn_classes: number of classes the student ishours_hw: hours of homework the student has

Selecting columns
We can select columns to keep in our dataset
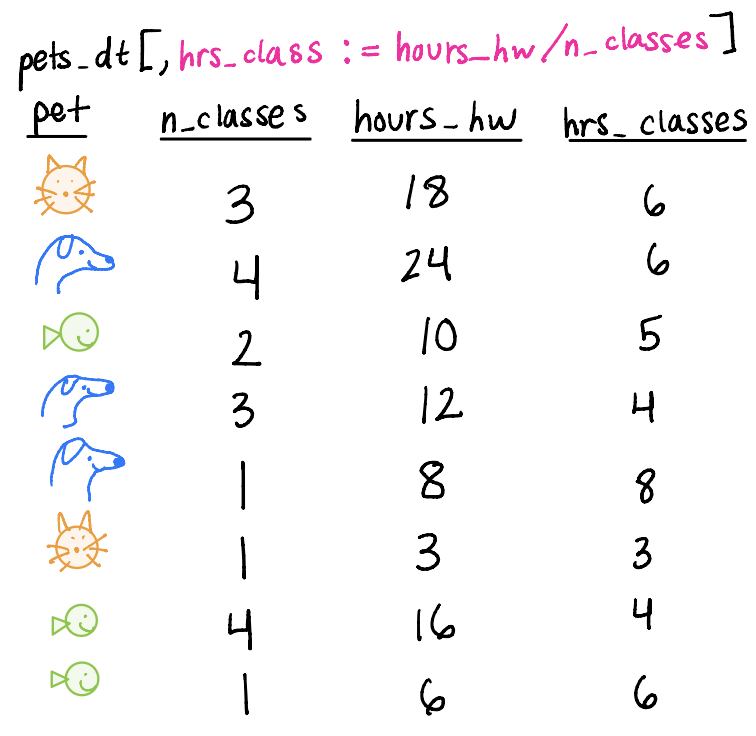
Mutating variables
We can add (mutate) variables to the dataset, keeping the same number of rows as before
Filtering rows
We can filter for particular type of row based on a logical statement

Arranging rows
We can arrange the dataset by a particular variable(s)

Grouping by and summarizing
We can create summary statistics (summarize) by particular groups (group_by)
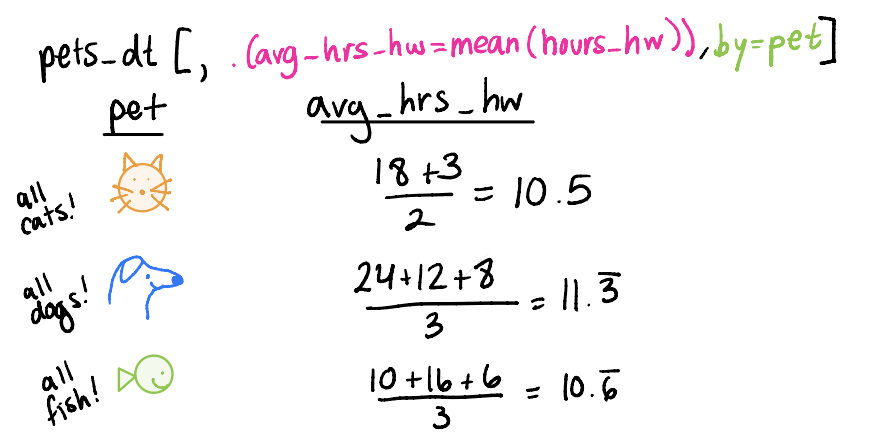
Let’s poll!

Joining datasets
We might also want to join two datasets, meaning we combine them based on the information in each respective one.
In data.table, joins are supported by the base syntax:

Note that joins are right joins by default in data.table!
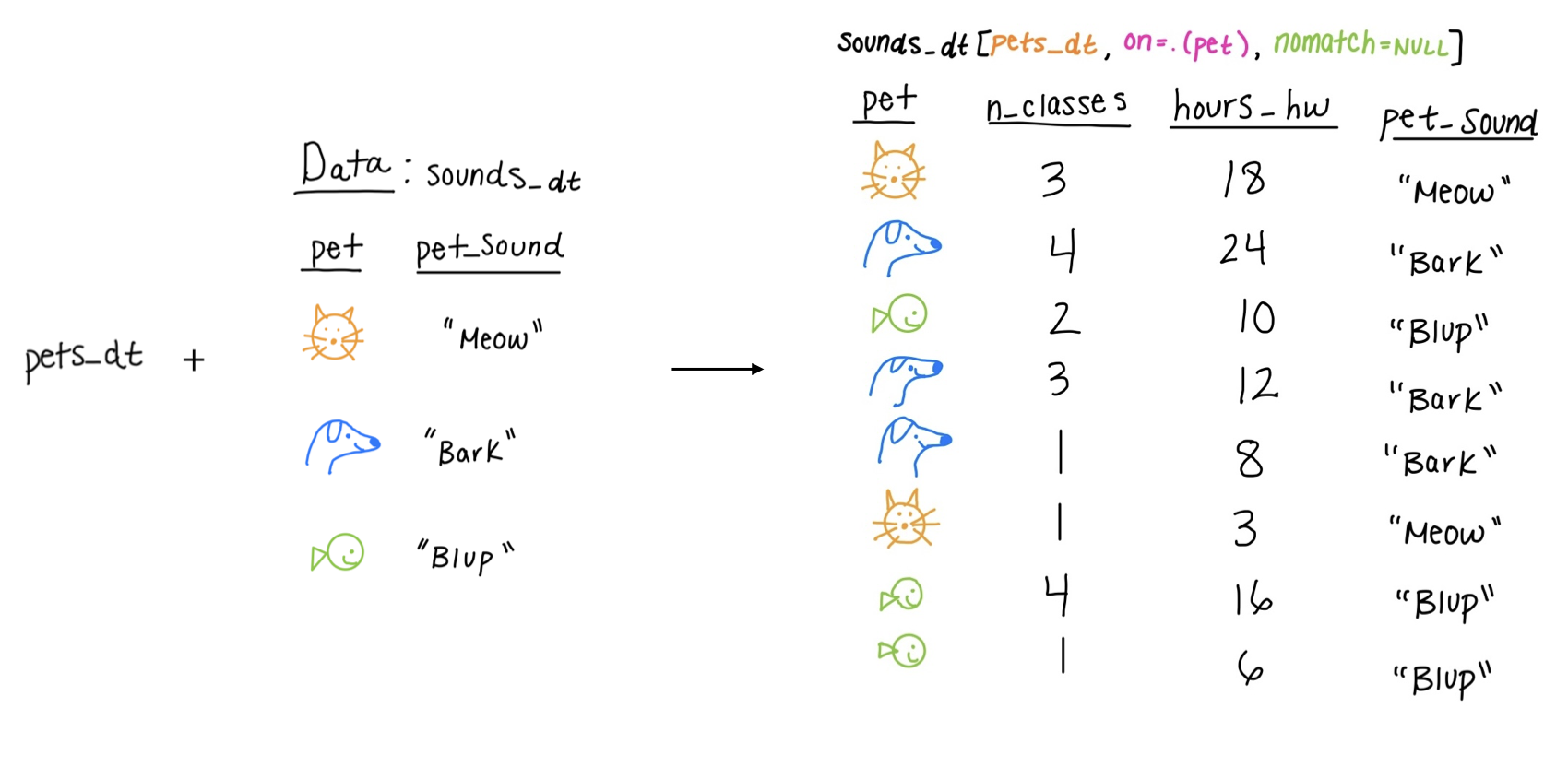
Equi joins
Equi joins: find common elements between the two datasets to combine
Inner Join Example:
Advanced join types
Non-Equi Joins: match rows based on comparison operators other than strict equality
- for example: \(>\), \(<\), \(\geq\), or \(\leq\) operators
Overlapping Joins: match rows based on overlapping ranges between elements
- see the
foverlaps()function
- see the
Rolling Joins: match rows based on the nearest value in a sorted column
- useful for time series data or for imputing missing values with nearest available data
Piping statements together
What if we wanted to pair multiple of the six main verbs together? For example, filter pets_dt to remove the cats and then arrange by hours_hw?
- We could save the filtered dataset to
no_cats_dtand then order that. - We could also pipe statements together!

Three equivalent ways to pipe
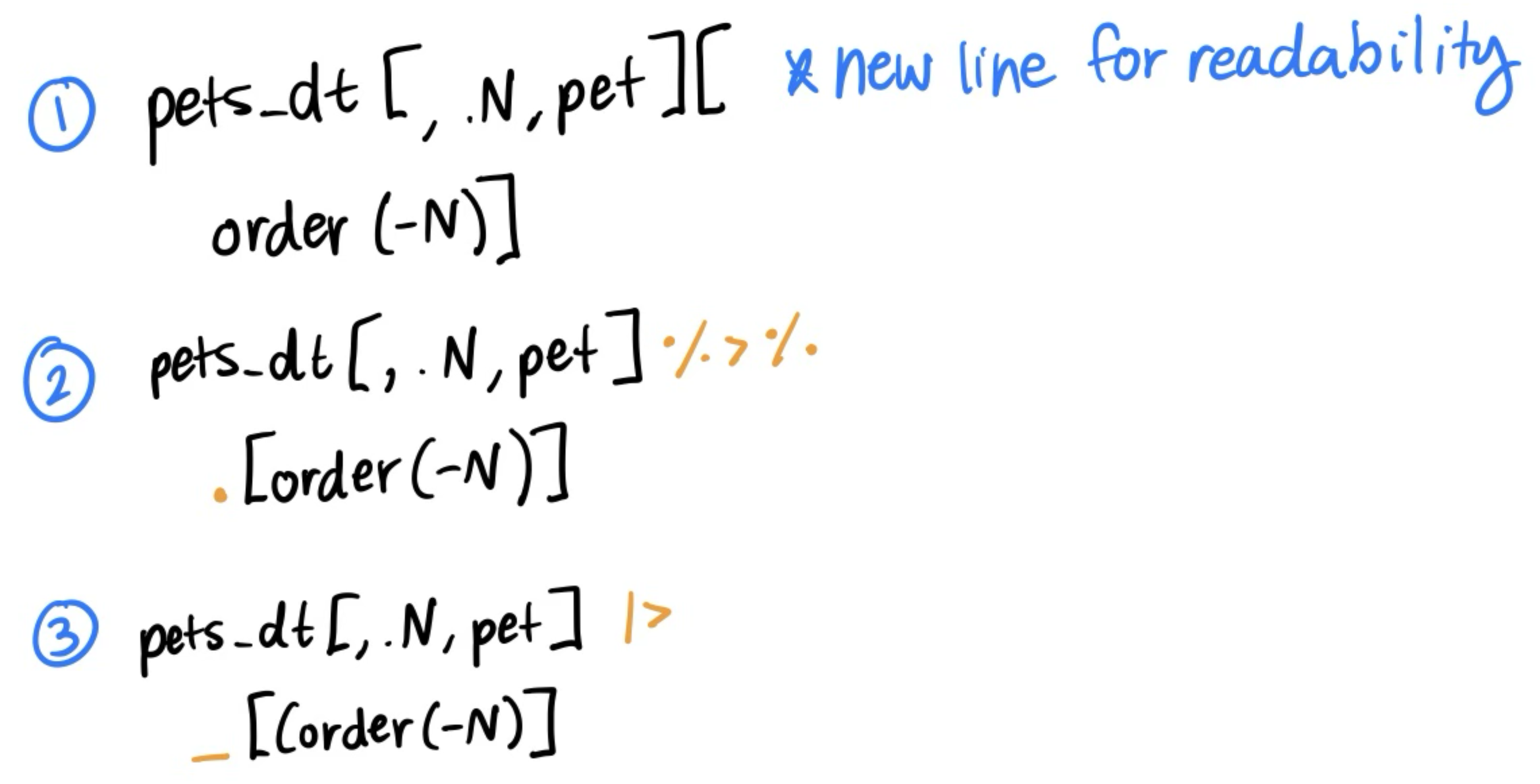
Reading and writing files
fread() and fwrite()
To (a) read a file into R or (b) save a file to home directory, fread() and fwrite() are efficient and support several file types.
Supports:
.csv, .tsv
Other delimited files (semicolon, colon, pipe)
Compressed files with .gz, .bz2 extensions
Nice arguments of fread()
nrow: number of rows to read in
pet n_classes hours_hw
<char> <int> <int>
1: cat 3 18
2: dog 4 24
3: fish 2 10skip: row number or string match to start reading from
Nice arguments of fread() (cont)
select: columns to keep
pet
<char>
1: cat
2: dog
3: fish
4: dog
5: dog
6: cat
7: fish
8: fishdrop: columns to remove
Bonus features
Rowwise data.table object creation
Convenient and readable for creating small datasets
- For instance, creating toy examples or summary tables
Visualizing by a grouping variable
Let’s say we are investigating the relationship between lot area and sale price and we want to see how it varies by driveway pavement.
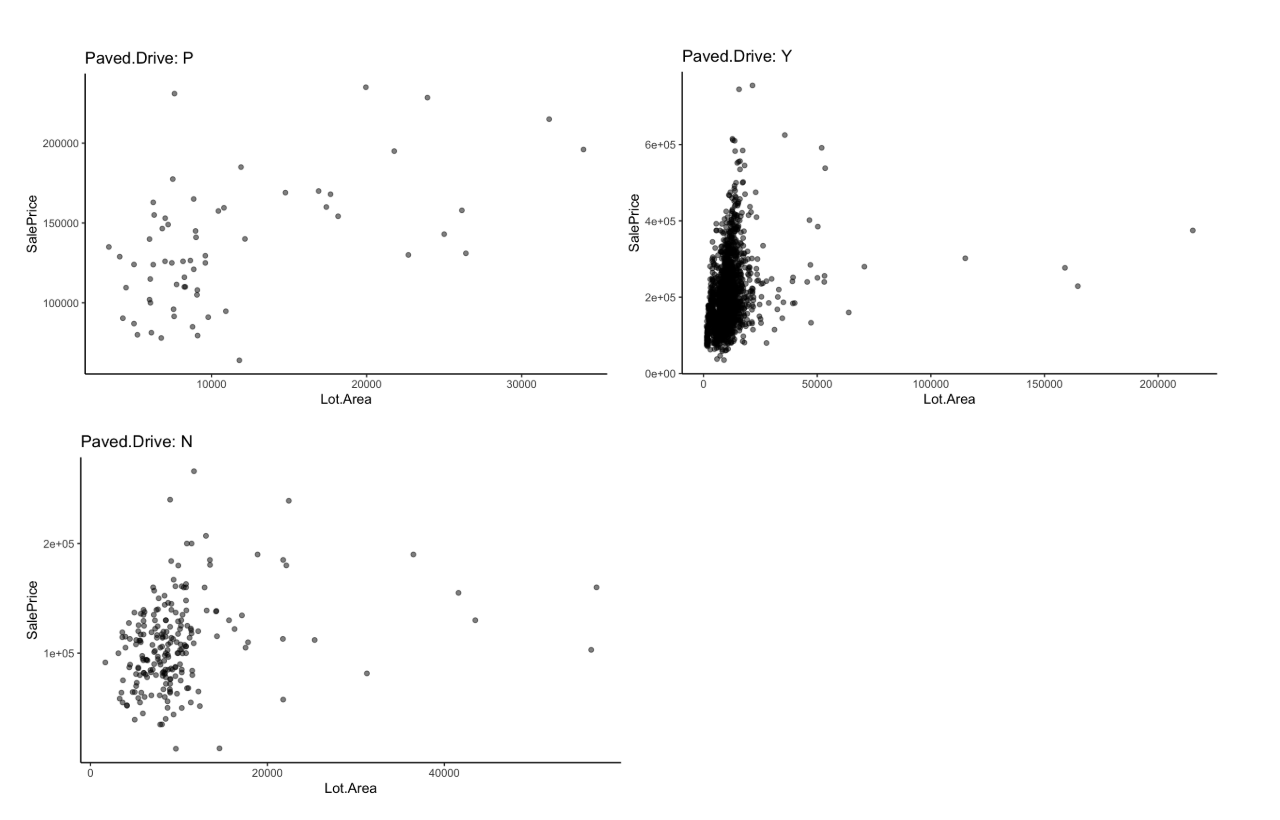
Modeling by a grouping variable
With data.table we can also create a distinct model for each group. For example, we can fit a linear model for sale price of homes in Iowa (SalePrice) for each neighborhood in the dataset:
We can then cross-compare the coefficient estimates and p-values for the covariates in our model to see how the differ between neighborhoods:
Old Town, Ames, Iowa:
| Characteristic | Beta | 95% CI | p-value |
|---|---|---|---|
| Lot.Area | 5.0 | 3.5, 6.4 | <0.001 |
| Bedroom.AbvGr | 6,895 | 1,299, 12,490 | 0.016 |
| Full.Bath | 17,015 | 5,888, 28,143 | 0.003 |
| Year.Built | -279,362 | -651,881, 93,157 | 0.14 |
| Yr.Sold | -268,748 | -625,248, 87,751 | 0.14 |
| Year.Built * Yr.Sold | 139 | -46, 325 | 0.14 |
| Abbreviation: CI = Confidence Interval | |||
Northridge Heights, Ames, Iowa:
| Characteristic | Beta | 95% CI | p-value |
|---|---|---|---|
| Lot.Area | 18 | 15, 22 | <0.001 |
| Bedroom.AbvGr | -14,055 | -30,965, 2,855 | 0.10 |
| Full.Bath | 5,483 | -36,505, 47,471 | 0.8 |
| Year.Built | -2,313,155 | -13,425,305, 8,798,994 | 0.7 |
| Yr.Sold | -2,315,556 | -13,410,216, 8,779,105 | 0.7 |
| Year.Built * Yr.Sold | 1,156 | -4,376, 6,689 | 0.7 |
| Abbreviation: CI = Confidence Interval | |||
Connection to tidyverse
Weighing the options
- Both
data.tableandtidyverseare great tools for wrangling data. Luckily, we are not confined to just one of them!1 - It all comes down to personal preference:
- Comfort with syntax
- Brevity of syntax
- Consistency with collaborators
- Computational efficiency 2

dtplyr
Perhaps you have some experience with data wrangling in the tidyverse, but you need the efficiency of data.table.
The dtplyr allows for data.table-like efficiency while still writing code in tidyverse, with a few small modifications.1
So, what do we mean when we say data.table is computationally efficient?

Interested in learning more about computational effiency?
This Stack Overflow thread explains a bit about memory usage
Visit this vignette to learn about keys and this vignette to learn about indexing.
- These features are super helpful when working with large data!
Thanks! Questions?
Post-Lecture Survey
