Rows: 3,504
Columns: 7
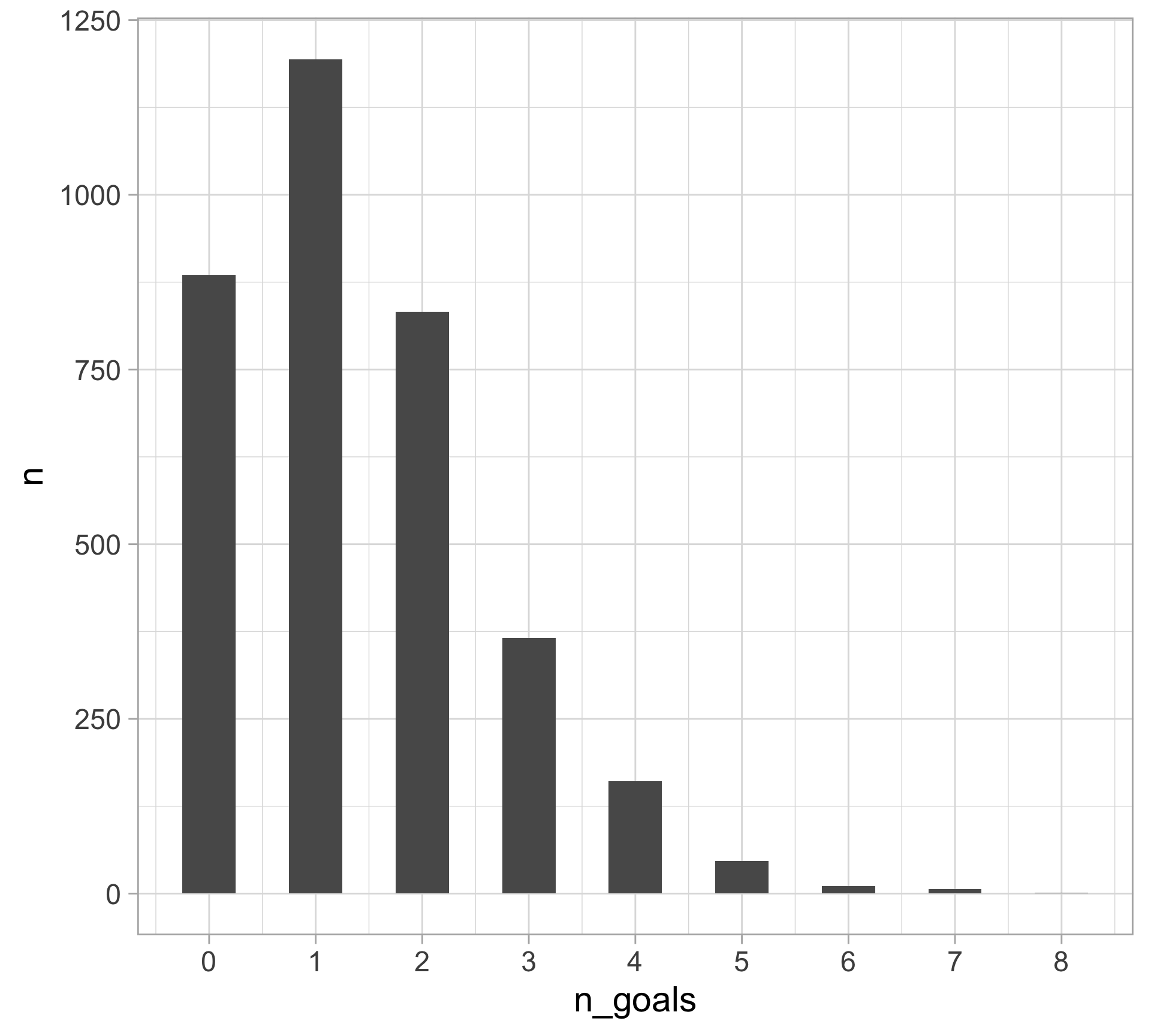
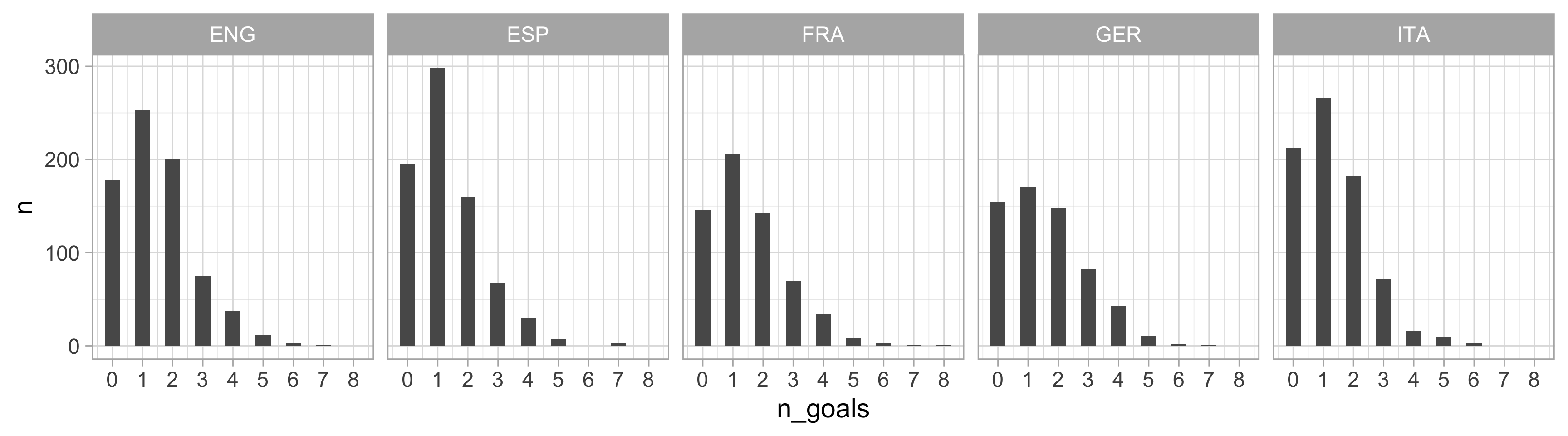
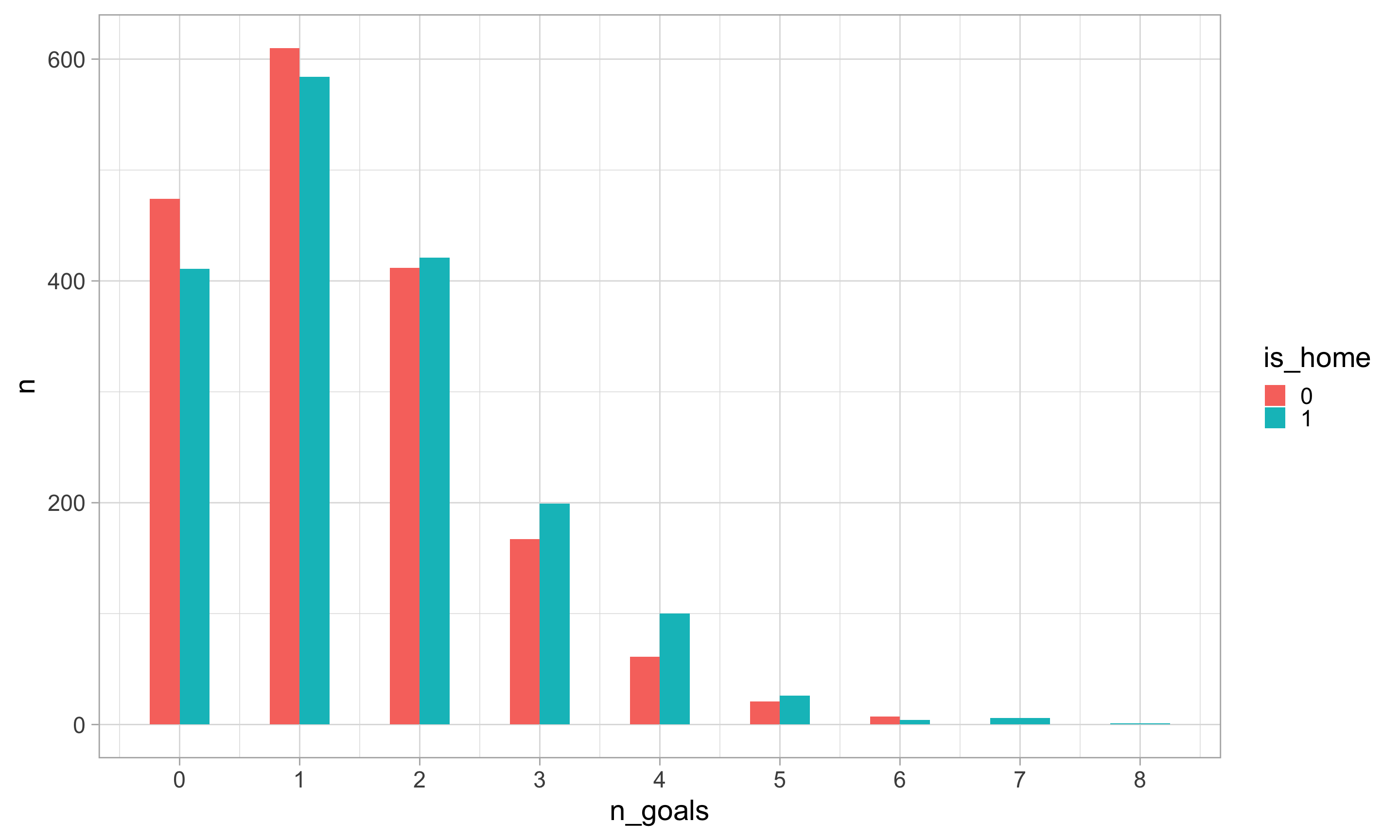
$ n_goals <dbl> 1, 0, 1, 0, 1, 2, 1, 2, 0, 1, 1, 2, 1, 3, 1, 2, 2, 4, 1, 2, 0…
$ xG <dbl> 2.4, 0.5, 0.3, 0.5, 1.3, 1.2, 2.3, 1.6, 1.0, 1.0, 0.5, 1.6, 0…
$ off_team <chr> "Manchester Utd", "Ipswich Town", "Newcastle Utd", "Everton",…
$ def_team <chr> "Fulham", "Liverpool", "Southampton", "Brighton", "Bournemout…
$ league <chr> "ENG", "ENG", "ENG", "ENG", "ENG", "ENG", "ENG", "ENG", "ENG"…
$ match_id <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18…
$ is_home <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…