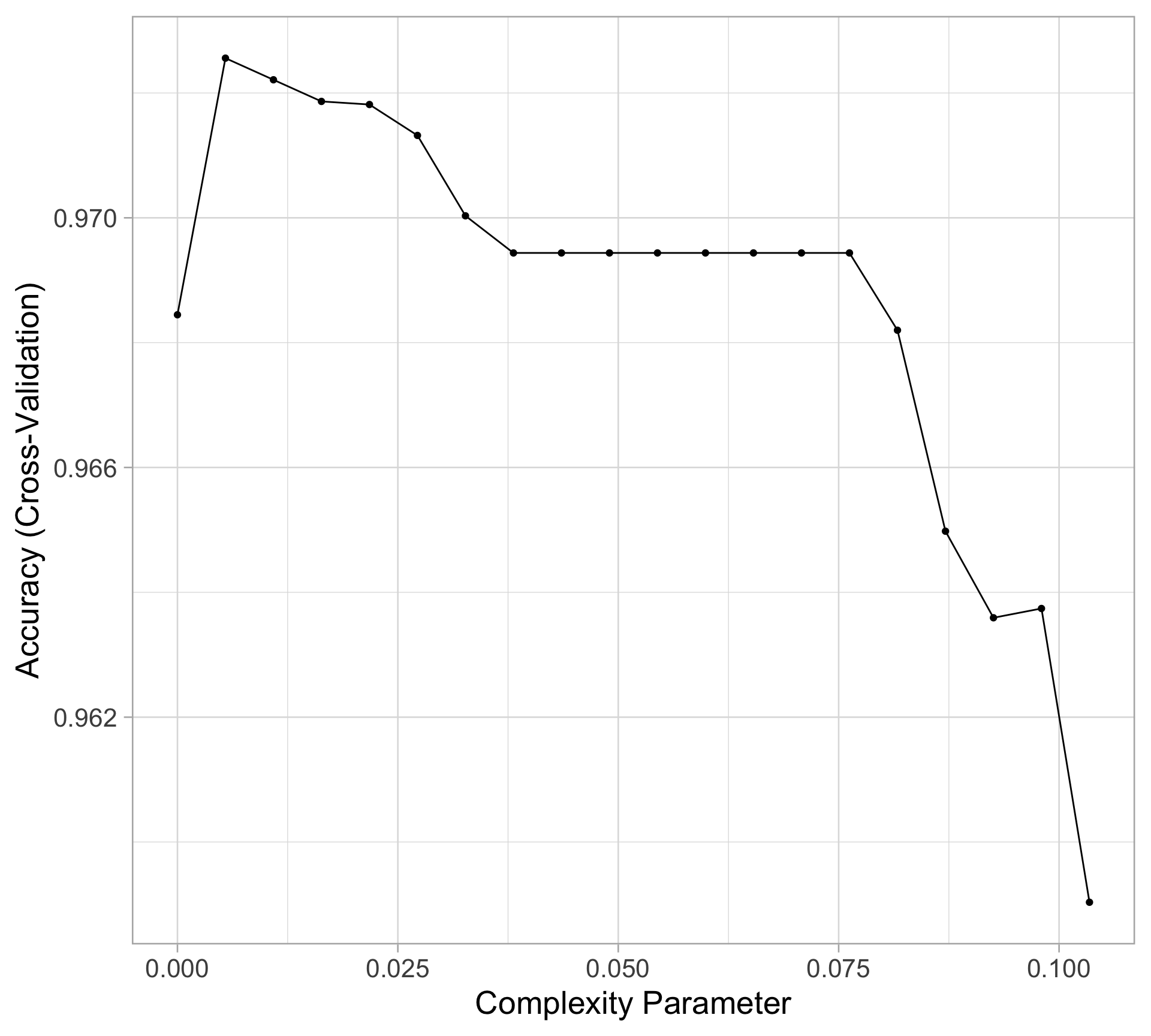
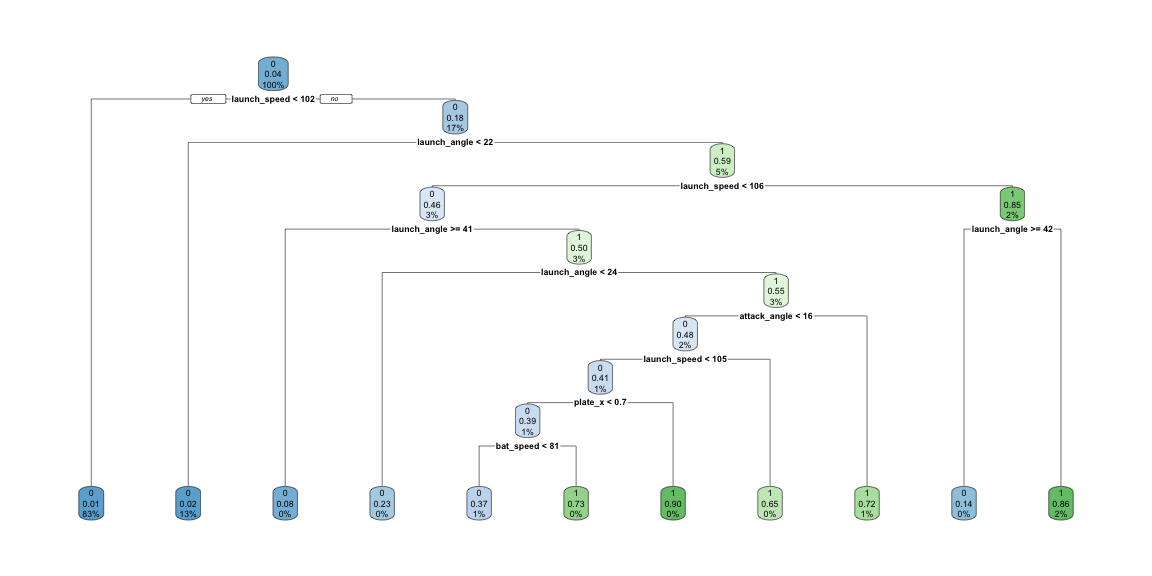
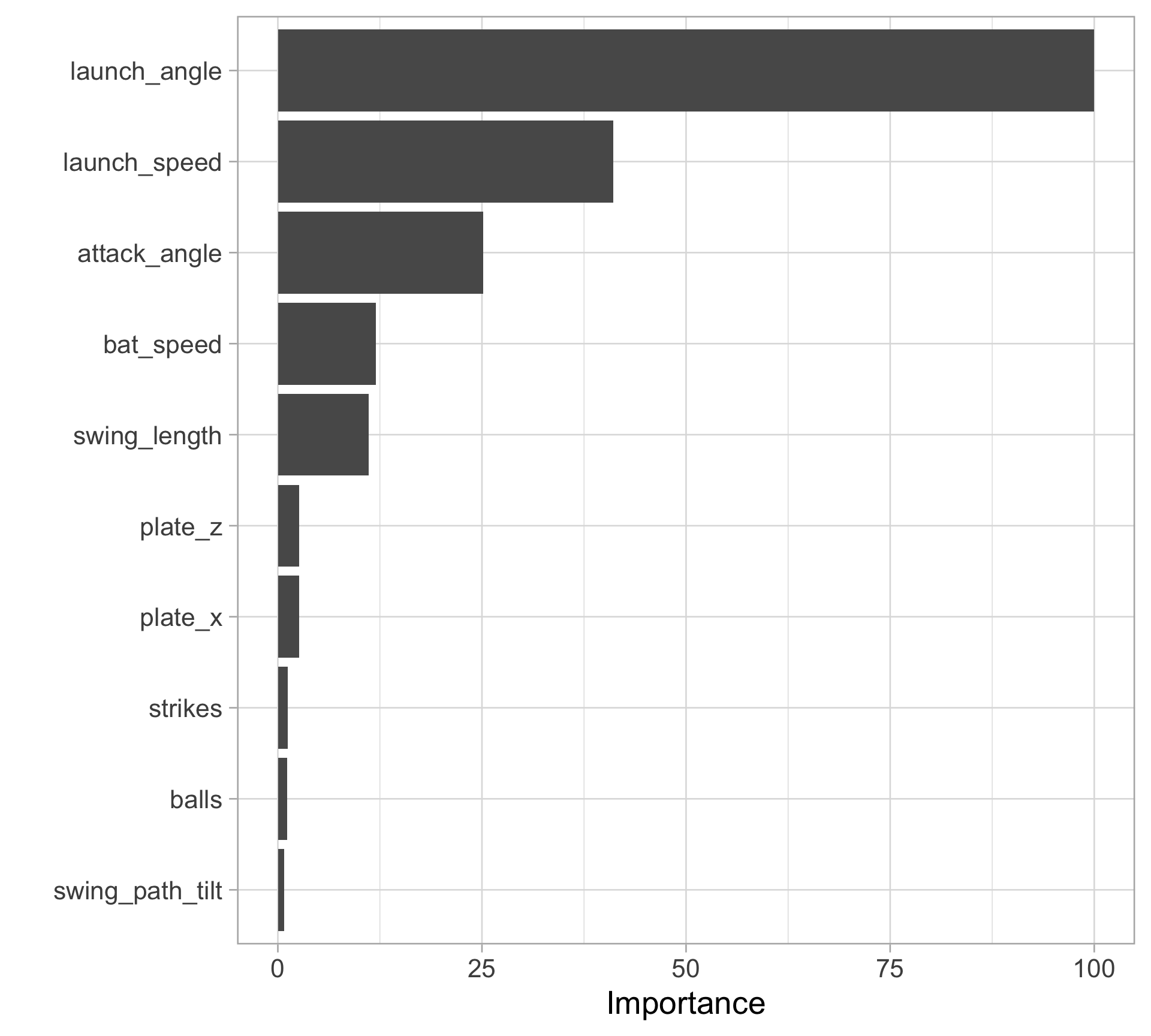
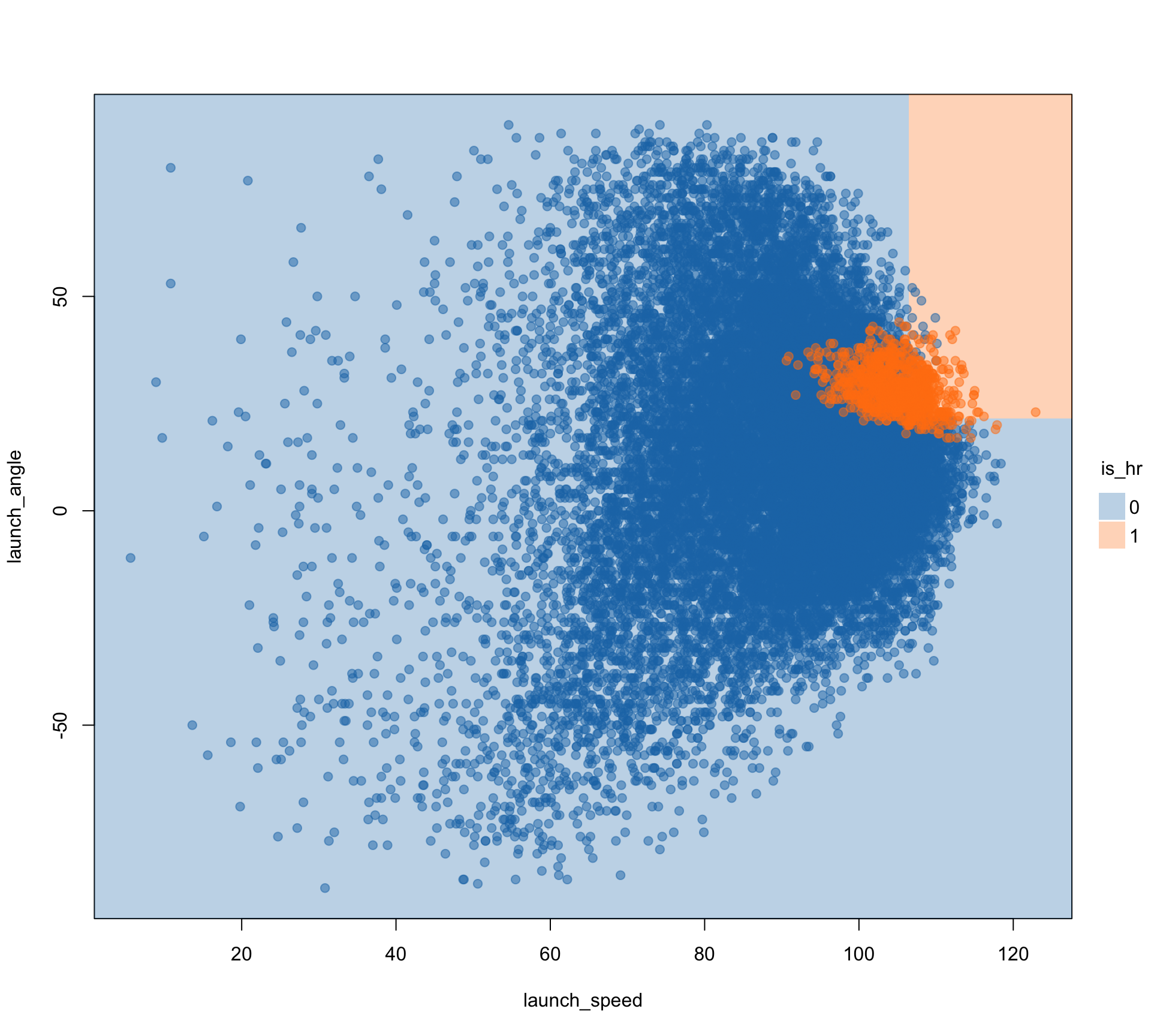
Rows: 20,188
Columns: 14
$ is_hr <fct> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ launch_angle <dbl> 33, -20, 36, 4, 20, 28, 27, 12, 8, -25, 16, 50, 68, 7,…
$ launch_speed <dbl> 91.3, 71.4, 103.4, 100.8, 102.3, 90.4, 90.4, 76.9, 101…
$ bat_speed <dbl> 76.3, 79.2, 72.3, 72.2, 67.8, 79.1, 74.7, 70.4, 74.3, …
$ swing_length <dbl> 8.2, 8.0, 6.4, 8.4, 5.9, 7.7, 8.2, 6.6, 7.0, 6.7, 6.6,…
$ attack_angle <dbl> 0.1394548, 22.2024920, 11.7628706, 3.3512694, -4.21284…
$ swing_path_tilt <dbl> 40.20981, 30.55564, 29.29605, 29.01934, 26.56705, 33.4…
$ plate_x <dbl> -0.45, 0.26, -0.04, -0.65, -0.40, -0.25, -0.23, -0.94,…
$ plate_z <dbl> 1.98, 2.02, 2.75, 1.81, 2.78, 2.08, 1.58, 2.14, 2.70, …
$ inning <dbl> 9, 9, 9, 8, 8, 8, 8, 7, 7, 6, 6, 6, 5, 5, 5, 5, 5, 5, …
$ balls <dbl> 0, 1, 3, 2, 0, 1, 1, 2, 3, 0, 0, 2, 0, 2, 0, 1, 0, 0, …
$ strikes <dbl> 2, 2, 2, 1, 0, 2, 2, 2, 2, 0, 0, 2, 0, 2, 0, 1, 1, 1, …
$ is_stand_left <dbl> 1, 0, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 1, 0, 1, …
$ is_throw_left <dbl> 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, …